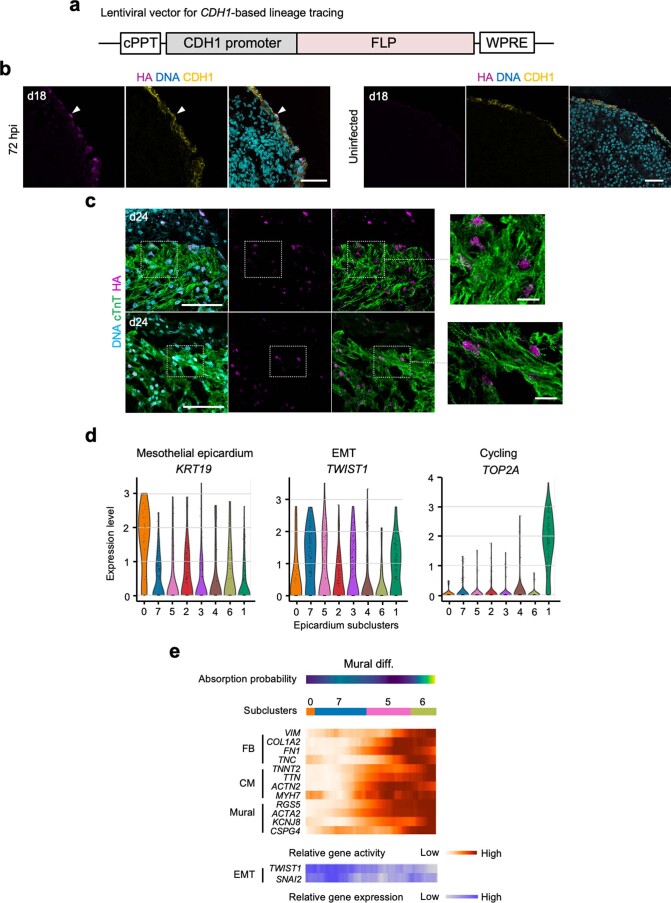
Extended Data Fig. 9. Analysis of epicardial cells and their derivatives by lineage tracing and multi-omic analysis.
(a) Schematic representation of the lentiviral vector used for lineage tracing of mesothelial epicardial cells in epicardioids derived from AAVS1-CAG-FRT-flanked STOP-mKate2-HA reporter hiPSCs, consisting of the sequence encoding flippase (FLP) driven by the CDH1 promoter. (b) (Left) Immunostaining for the HA-tag and CDH1 in epicardioids at day 18, 72 h after infection with the CDH1-FLP lentivirus. The arrowhead shows an exemplary HA-tag+ CDH1+ epicardial cell at the outer layer. (Right) Corresponding uninfected negative control. Images are representative of 3 independent differentiations. Scale bars = 50 µm. (c) Immunostaining for the HA-tag and cardiac troponin T (cTnT) in infected organoids at day 24, showing HA-labeled cardiomyocytes (CMs) close to the epicardium (representative of 5 independent differentiations). Scale bars = 50 µm. Insets show exemplary labeled CMs at higher magnification; scale bars = 10 µm. (d) Violin plots showing the expression levels of markers of mesothelial epicardium (KRT19), EMT (TWIST1), and cycling cells (TOP2A) in epicardial metacell subclusters obtained from analysis of epicardioids by paired scRNA-seq and scATAC-seq. (e) Heatmap showing the relative gene activity of FB (VIM, TNC, FN1, COL1A2), CM (TNNT2, TTN, ACTN2, MYH7) and mural (RGS5, KCNJ8, ACTA2, CSPG4) lineage markers (orange) and the relative gene expression of EMT markers (TWIST1, SNAI2) (blue) along an alternative mural differentiation trajectory as the one presented in Fig. 5g. CM: cardiomyocyte; FB: fibroblast; EPDC: epicardium-derived cell; EMT: epithelial-to-mesenchymal transition.