Fig. 3. Effect of cholesterol on membrane bending modulus.
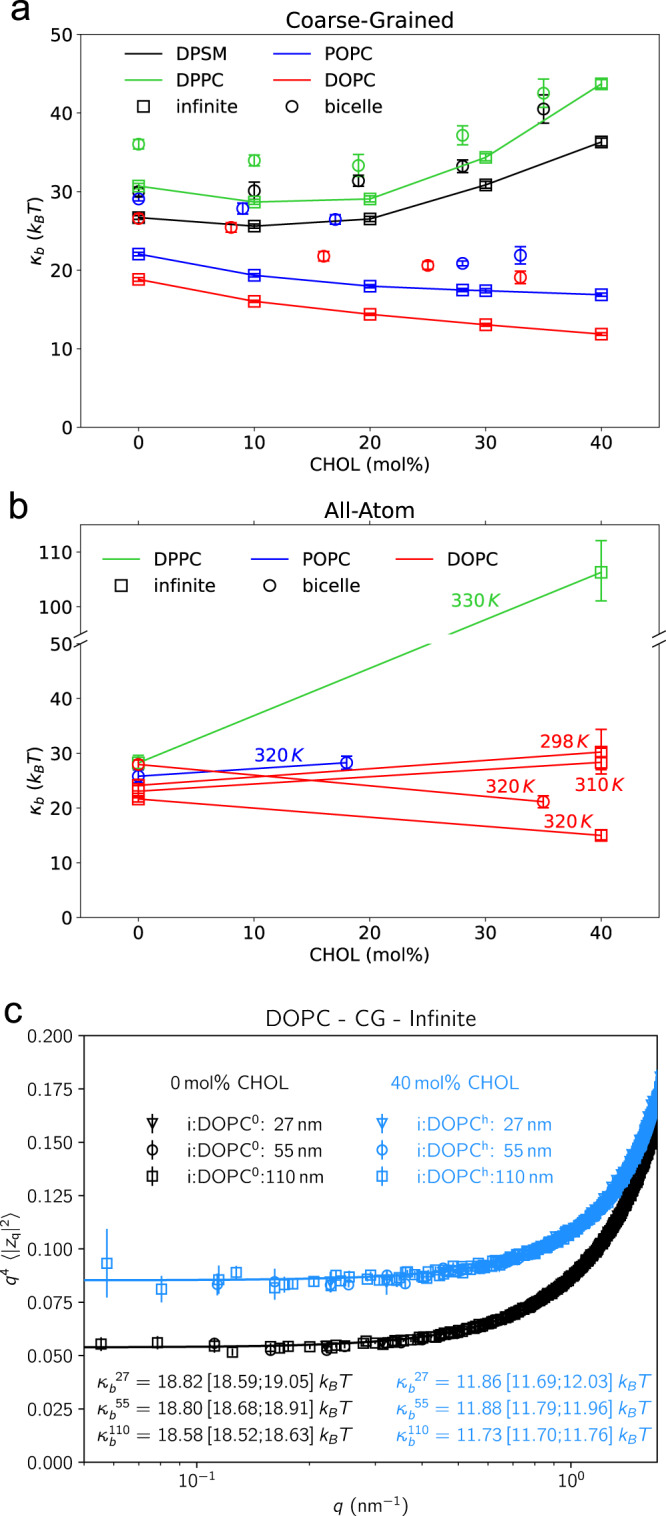
Membrane bending moduli were determined from CG MD simulations (a) and from all-atom MD simulations (b) for different system setups (bicelles as circles and infinite, periodic bilayers as squares). The bending moduli were calculated using undulation analysis (for infinite systems) and the local fluctuation method (for bicelles). Panel c shows exemplarily the spectrum for DOPC bilayer systems with 0% and 40% cholesterol for different lateral box sizes (27 nm, 55 nm, and 110 nm; are the amplitudes of the Fourier-transformed membrane surface). Displayed are mean values and errors as 95% confidence intervals employing parametric bootstrapping (N = 50,000 statistically independent samples) assuming Gaussian distributions of the mode-dependent amplitudes78 (infinite bilayers) or standard errors of the mean employing block averaging (at least N = 12 independent blocks; bicelles).
