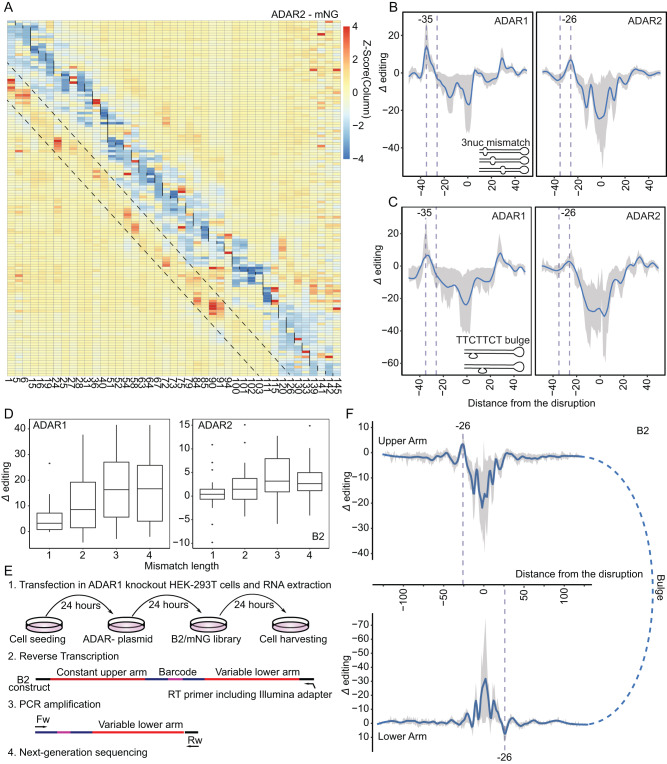
Fig. 2. ADAR2-mediated editing is induced at a constant interval of 26 bp upstream from structural disruptions.
A Heatmap of a 3-nucleotide mismatch running from 5’ to 3’ throughout the double-stranded RNA. Rows represent structurally disrupted constructs at specific positions, columns represent adenosine positions, and Δ editing is color-coded after Z-score transformation (mNG series). Vertical black lines mark the 3 nt mismatch location, and dashed lines highlight ADAR2-mediated editing increase upstream. B ADAR1- and ADAR2-mediated editing offsets based on the subset of 3-nucleotide mismatch running throughout the mNG and B2 sequences. Mismatches differentially located in each construct get centered at 0 on the x-axis. The Δ editing level on the y-axis represents the change of the editing level of an adenosine, normalized to the perfect double-stranded construct. Fitted curves depict the LOESS fit (blue-colored) of Δ editing with a span of 0.05 and the gray-shaded region spans the 25th percentile and 75th percentile values of Δ editing per distance. Only adenosine positions, which have greater than 1% in editing on the perfect double-stranded construct, were considered. Vertical dashed lines are placed at −26 and −35. C Subset of TTCTTCT bulges running throughout the mNG and B2 sequences. Data is shown as the LOESS fit curve (blue-colored) of Δ editing with a span of 0.11 and the gray-shaded region spans the 25th percentile and 75th percentile. D The mismatch size affects ADAR1- and ADAR2-mediated editing on adenosines located at −35 and −26 downstream from the mismatch, respectively. Data is visualized via box-and-whisker plots, with the central line denoting the median, box edges representing the interquartile range (from the 25th to the 75th percentile), and whiskers indicating the 1.5 times interquartile range. E Library preparation: RNA was extracted, the B2 variable lower arm and barcode were reverse transcribed, and subsequently PCR amplification and sequencing using Novaseq 6000 platform with a 300 bp kit were performed. F Depiction of the subset of 3-nucleotide mismatch running throughout the stem (B2) in ADAR1-knockout HEK293T cells overexpressing ADAR2. Constant and variable arms are illustrated under each other, and nucleotide locations are aligned. Data is shown as Fig. 2B.