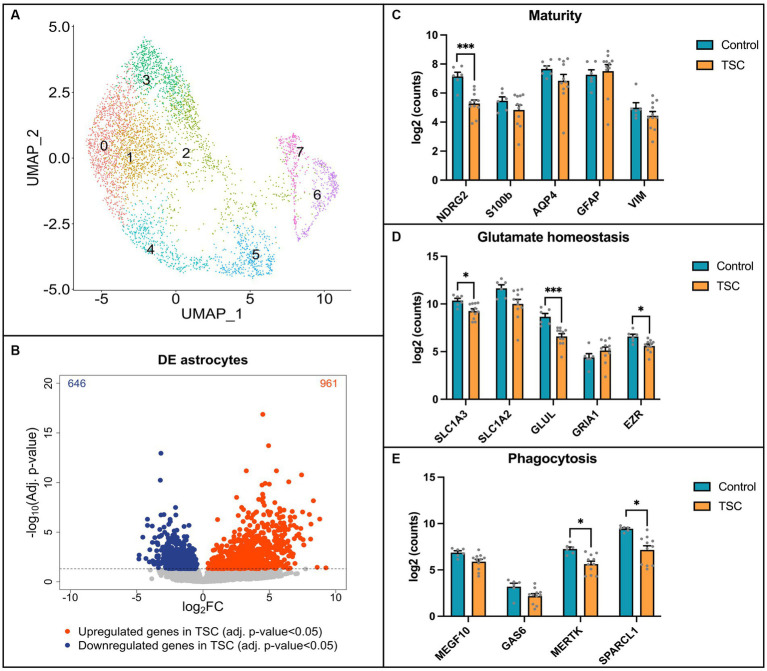
Figure 4.
Single-nuclei RNA sequencing reveals alterations in key pathways in TSC astrocytes. (A) Uniform Manifold Approximation and Projection (UMAP) plot displaying the clustering of astrocytes. Eight distinct clusters were identified and found in both control (n = 6) and Tuberous Sclerosis Complex (TSC) (n = 11) tissue. (B) Volcano plot illustrating the differential gene expression analysis based on control (n = 6) and TSC (n = 11) samples. A total of 646 downregulated genes and 961 upregulated genes were identified in TSC when compared to control samples. The analysis was performed using DESeq2 after pseudobulk of the count values. Genes with a value of p less than 0.05 were considered statistically significant and are depicted in blue (downregulated) or red (upregulated) dots. (C) Box plot representation of log2-transformed count values for genes associated with astrocyte maturity. Notably, the expression of NDRG2 was significantly downregulated in TSC compared to control samples. (D) Box plot depicting the expression of genes related to glutamate homeostasis. In TSC, there was a downregulation of SLC1A3 (EAAT1), GLUL (GS), and EZR indicating alterations in glutamate regulation and metabolism. (E) Box plots showing the expression levels of genes involved in astrocyte phagocytosis. The expression of MERTK and SPARCL1 was significantly downregulated in TSC, suggesting a dysregulation of astrocyte-mediated phagocytic processes. Statistical significance is denoted as * (p < 0.05) or *** (p < 0.001) in all box plots as determined by DESeq2, indicating significant differences between control and TSC samples. Samples were corrected for multiple comparison (Benjamini–Hochberg).