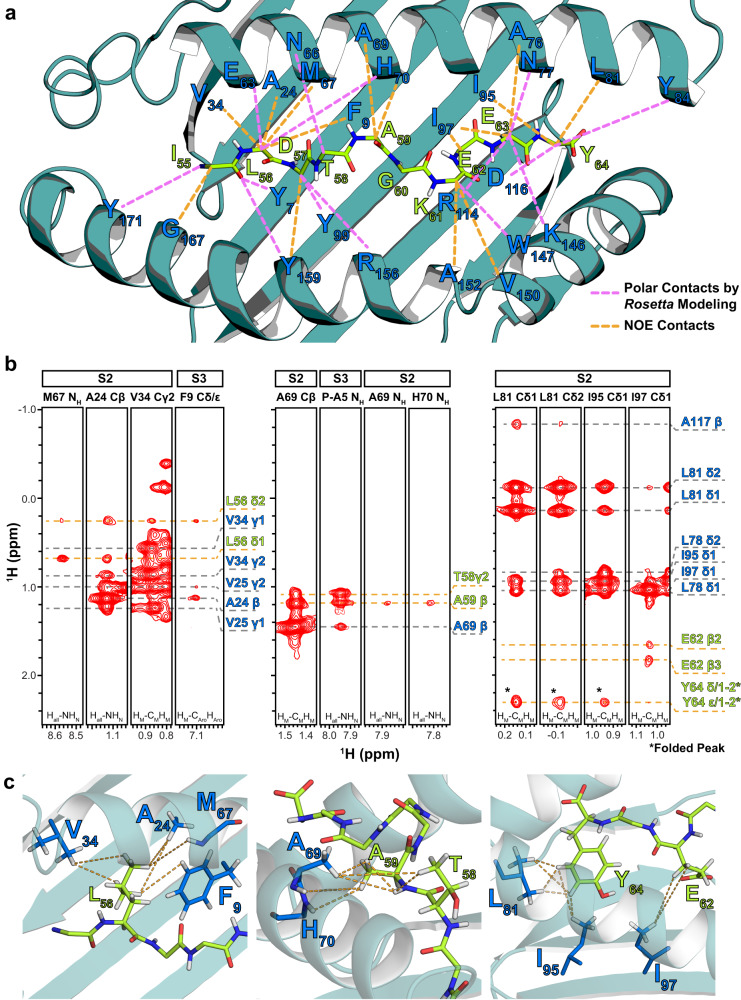
Fig. 2. Intermolecular NOE contacts observed between the HLA-A*01:01 groove and NRASQ61K.
a Intermolecular NOEs observed between HLA-A*01:01 residues and NRASQ61K residues (dotted orange lines) in 3D SOFAST NOESY experiments are shown with HLA-A*01:01 groove (dark green) and peptide residues (green) labeled. Intermolecular polar contacts not observed experimentally, but observed in the final structure following high-resolution refinement using the Rosetta FlexPepDock ab-initio protocol are also shown (dotted purple lines). b Representative 1HM-1HM NOESY strips corresponding to intra- (gray dotted lines) and inter-molecular (orange dotted lines) NOE contacts, shown for regions spanning the HLA-A*01:01 groove (A/B pocket – left, B/C pocket – center, and E/F pocket – right). The top of the strip is labeled with the name of the sample used (S2 or S3) and the residue/atom name corresponding to the strip. The bottom of the strip is labeled with the NMR experiment where the NOE strip was obtained. Atoms corresponding to HLA-A*01:01 are labeled blue, while atoms corresponding to NRASQ61K are labeled green. NOE cross-peaks that are folded due to sweep width restrictions are indicated with an asterisk (*). c View of intermolecular NOE contacts (dotted lines) observed in panel B with HLA-A*01:01 (dark green) and peptide (light green) side chains shown as sticks. Our final refined structure of the complex (PDB ID 6MPP) was used to generate structure views.