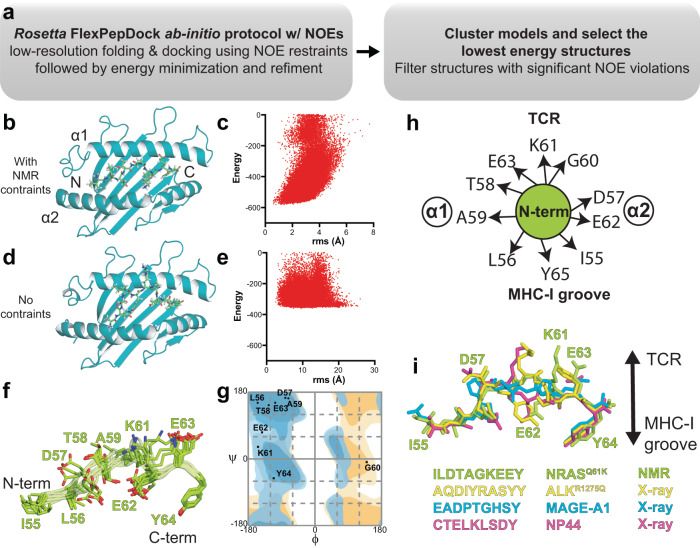
Fig. 3. Molecular surface features of the NRASQ61K/HLA-A*01:01 groove for TCR recognition.
a Flow chart of the combined NMR and Rosetta FlexPepDock ab-initio protocol. b, d The Rosetta FlexPepDock ab-initio protocol was performed b with and d without NMR restraints. The HLA-A*01:01 groove is shown in dark green and NRASQ61K is shown in light green. The peptide cannot properly dock into the HLA-A*01:01 groove without NMR restraints due to geometric restrictions imposed by the MHC-I pockets. c, e Energy landscape plots showing energy (Rosetta energy units) versus backbone heavy atom root mean squared deviation (r.m.s., Å) to the lowest energy structure are shown for 50,000 models of FlexPepDock ab-initio protocol performed with c NOE-derived NMR constraints (Supplementary Table 2) and e without NMR constraints. f Overlay of the ten lowest energy NRASQ61K conformations in complex with HLA-A*01:01/β2m (not shown for clarity). g Ramachandran plot showing the φ/ψ angles of each residue of NRASQ61K peptide bound to HLA-A*01:01. The φ/ψ were computed on the peptide in the lowest energy structure HLA-A*01:01 bound NRASQ61K structure using the RAMPAGE server. Blue – general amino acid favored/allowed. Orange – glycine favored/allowed. h Cartoon schematic of the side chain orientation of the NRASQ61K (bound to HLA-A*01:01) displayed to TCRs as viewed from the N-terminal end relative to the MHC-I α1/α2 helices. i Comparison of the conformation and surface features of NRASQ61K (PDB ID 6MPP) displayed by HLA-A*01:01 to TCRs with peptides taken from X-ray structures of other known HLA-A*01:01-restricted epitopic peptides (ALKR1275Q - PDB ID 6AT9, MAGE-A1 - PDB ID 3BO8, NP44 - PDB ID 4NQV).