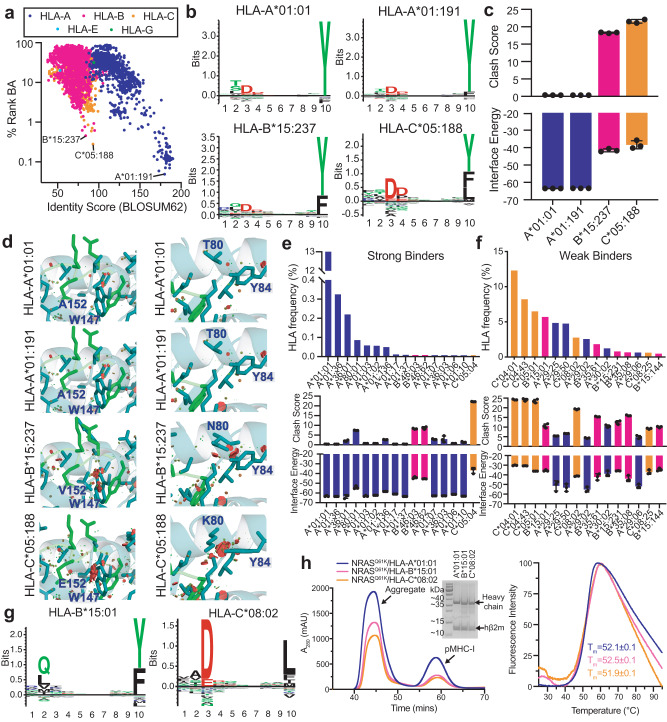
Fig. 7. In silico and in vitro evaluation of the HLA binding repertoire of NRASQ61K.
a In silico prediction of NRASQ61K (ILDTAGKEEY) binding to 10,386 different HLA molecules with netMHCpan-4.1. The % rank binding affinity (BA) from netMHCpan-4.1 is compared to the BLOSUM62 identity score for all HLA alleles relative to HLA-A*01:01, calculated for HLA groove residues. The top predicted NRASQ61K binders for each HLA-A-, B-, and C- class are noted. b Decamer peptide binding motifs (generated by Seq2Logo) for the top predicted NRASQ61K binders for each HLA-A-, B-, and C- class. c Interface energy (REU) obtained from RosettaMHC models of NRASQ61K with the top predicted binders for each HLA-A-, B-, and C- classes and their MolProbity clash score. Our solved NMR structure (PDB ID 6MPP) was used as the template for modeling. Data are mean ± SD for n = 3 independent experiments. d Zoom view of RosettaMHC models of NRASQ61K with the top predicted binders for each HLA-A-, B-, and C- class. HLA residues within 3.5 Å of the peptide are shown. Van der Waals overlaps or steric clashes are represented with red/green discs. e and f Top: HLA frequency (%, obtained from the Allele Frequency Net Database for all populations) for the top predicted e strong and f weak NRASQ61K binders. Bottom: Interface energy (REU) obtained RosettaMHC models of NRASQ61K with the most frequently represented strong and weak binders in the global population and their MolProbity clash scores. PDB ID 6MPP was used as the template for modeling. Data are mean ± SD for n = 3 independent experiments. g Decamer peptide binding motifs (generated by Seq2Logo) for HLA-B*15:01 and HLA-C*08:02, which are predicted as weak binders of NRASQ61K. h Left: SEC traces of in vitro refoldings of NRASQ61K with HLA-A*01:01/hβ2m, HLA-B*15:01/hβ2m, and HLA-C*08:02/hβ2m. The SDS-PAGE gel of the purified pMHC-I complexes is also shown. Right: DSF experiments performed on the purified pMHC-I complexes with fitted thermal stability (Tm, °C) shown. Data are mean ± SD for n = 3 technical replicates.