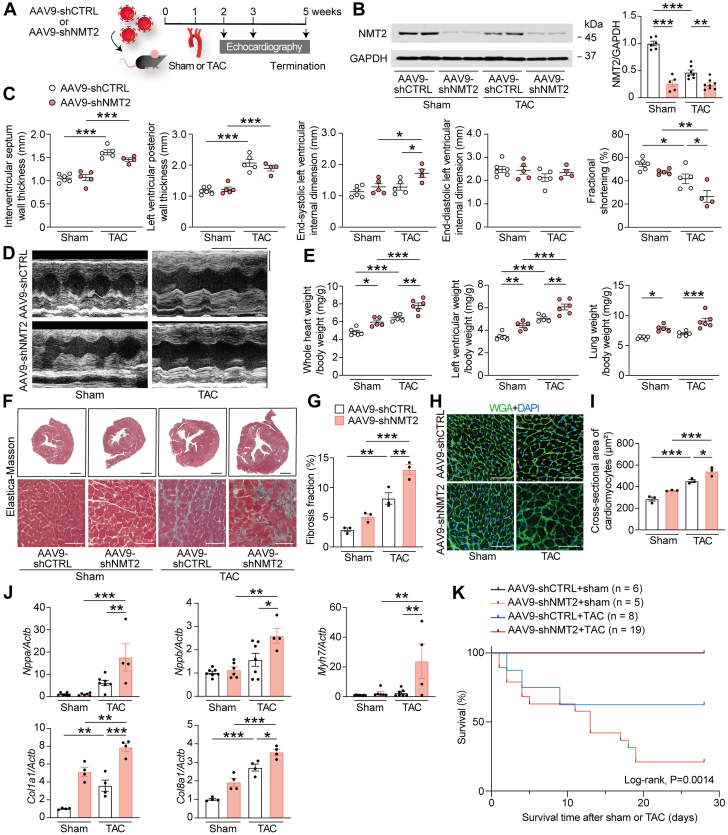
Figure 2.
NMT2 Knockdown Is Maladaptive in a Mouse Model of Pathological Cardiac Hypertrophy and Failure
(A) A schematic diagram of the experimental design. The mice at the age of 6 weeks were injected with 1.0 × 1011 genome-containing units of adeno-associated virus 9 (AAV9) encoding short hairpin RNA (shRNA) target to NMT2 (AAV9-shNTM2) or LacZ as a control (AAV9-shCTRL). One week later, TAC surgery using a 27-gauge blunt needle or sham procedure was performed. (B) Immunoblot analysis for NMT2 in the heart. The protein extracts from the left ventricles in mice receiving either AAV9-shCTRL or AAV9-shNMT2 at 4 weeks after sham or TAC operation were immunoblotted with the indicated antibodies. GAPDH was used as the loading control. The densitometric analysis is shown in the graphs (n = 5-8 in each group). (C) Echocardiographic assessment. Sham-operated AAV9-shCTRL–injected mice (n = 6), sham-operated AAV9-shNMT2–injected mice (n = 5), TAC-operated AAV9-shCTRL–injected mice (n = 5), and TAC-operated AAV9-shNMT2–injected mice (n = 4). (D) Representative images of M-mode echocardiograms. Scale bars = 0.2 seconds and 2 mm, respectively. (E) Physiological parameters. (F) Elastica-Masson–stained sections of the left ventricles. Scale bar = 1 mm (top) and 20 μm (bottom). (G) Quantitative analysis for fibrosis fraction in Elastica-Masson–stained sections (n = 3 in each). (H) Wheat germ agglutinin (WGA)-stained sections from the left ventricles. The plasma membrane and nucleus are stained with WGA (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue), respectively. Scale bar = 20 μm. (I) Quantification of the cross-sectional area of cardiomyocytes in WGA-stained sections (n = 3 in each). (J) Messenger RNA expressions of Nppa, Nppb, Myh7, Col1a1, and Col8a1. Actb was used for normalization. The average value for sham-operated AAV9-shCTRL–injected mice was set equal to 1 (n = 4-6). (K) Kaplan-Meier survival curve in AAV9-shCTRL– or AAV9-shNMT2–injected mice after sham or TAC surgery. The numbers of mice and P value by the log-rank test are presented. All data are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 by 1-way analysis of variance with Tukey’s post hoc analysis. Abbreviations as in Figure 1.