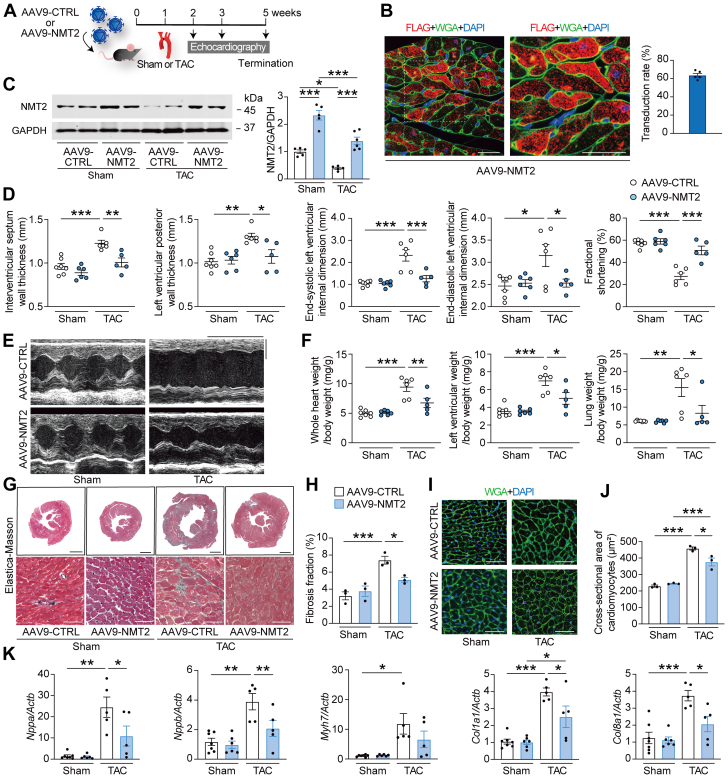
Figure 8.
NMT2 Gene Transfer Specific to the Heart Prevents Cardiac Pathological Hypertrophy and Remodeling in a Murine Model of Heart Failure
(A) Experimental design. AAV9 encoding mouse NMT2 with a FLAG-tag (AAV9-NMT2) with 1.0 × 1011 genome-containing units were injected in mice at the age of 6 weeks. AAV9 encoding mouse LacZ was used as a control (AAV9-CTRL). One week later, TAC surgery using a 28-gauge blunt needle or sham procedure was performed. (B) Representative images of immunohistochemistry for FLAG (red) and wheat germ agglutinin (WGA) (green) with DAPI (blue) from left ventricular tissue sections 5 weeks after AAV9-NMT2 injections. Boxed areas are highlighted in the right panel. Scale bars = 20 μm. The transduction rate of AAV9-NMT2 in the left ventricles is presented in the graph according to the ratio of cardiomyocytes with FLAG-positives (n = 5). (C) Immunoblot analysis for NMT2 in the heart. (D) Echocardiographic parameters. Sham-operated AAV9-CTRL–injected mice (n = 7), sham-operated AAV9-NMT2–injected mice (n = 6), TAC-operated AAV9-CTRL–injected mice (n = 6), and TAC-operated AAV9-NMT2–injected mice (n = 5). (E) Representative images of M-mode echocardiograms. Scale bars = 0.2 seconds and 2 mm, respectively. (F) Physiological parameters. (G) Elastica-Masson–stained sections of the left ventricles. Scale bar = 1 mm (top) and 20 μm (bottom). (H) Quantitative analysis of fibrosis fraction in Elastica-Masson–stained sections (n = 3 in each). (I) Representative images of WGA (green)- and DAPI (blue)-stained sections. Scale bar = 20 μm. (J) Quantitative analysis for the cross-sectional area of cardiomyocytes from WGA-stained sections. More than 100 cardiomyocytes were analyzed (n = 3 in each). (K) Messenger RNA expression of Nppa, Nppb, Myh7, Col1a1, and Col8a1 from the left ventricles by reverse transcription–quantitative polymerase chain reaction. Actb was used as the loading control. The average value for sham-operated AAV9-CTRL–injected mice was set equal to 1 (n = 5-7). All data are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 by 1-way analysis of variance with Tukey’s post hoc analysis. Abbreviations as in Figures 1 and 2.