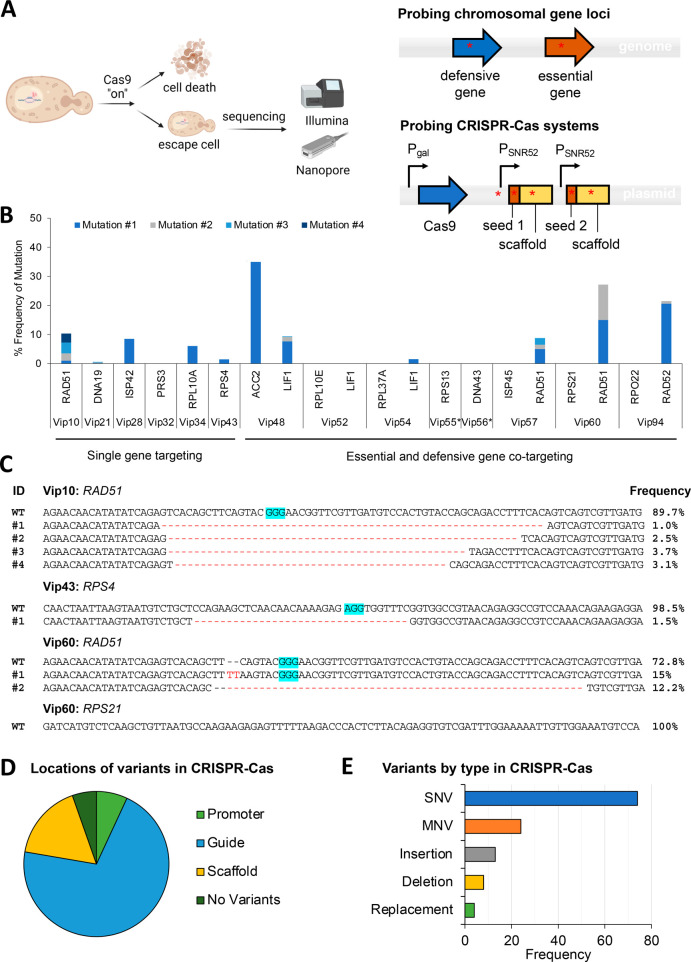
Figure 4.
Identification of mutations from representative single- and co-targeted strains that escape from the CRISPR-Cas antifungal treatment. (A) Experimental workflow for sequencing. (B) Genetic mutation frequency of the targeted chromosomal genes. Different colored stacked bars represent different mutations presented as mutation ID numbers in Figures 4C and S4. Reads for defensive genes of Vip55 and Vip56 failed QC. (C) Mutated sequences for the representative strains Vip10, Vip43, and Vip60. PAM sequences are highlighted in blue. Single targeted strains show low levels of mutation with the exception of the defensive genes, in particular RAD51. As seen in the sequence alignments provided for Vip10, Vip43, and Vip60, mutants are dominated by large (>20 bp) deletions, although mutation frequency is low and heterogeneous. Mutated sequences for other characterized strains were presented in Figure S4. (D) Locations of mutations in the gRNA cassettes on extracted gRNA plasmids. (E) Variants by type in gRNA cassettes.