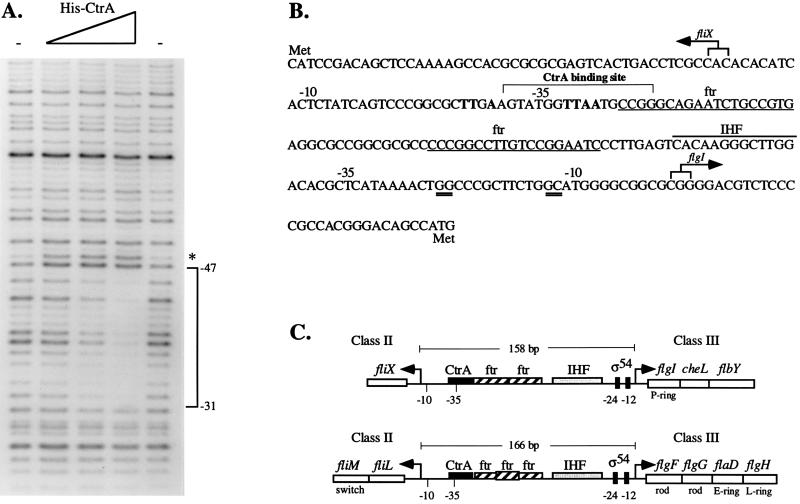
FIG. 7.
(A) Footprinting analysis of His-CtrA at the fliX promoter. An end-labeled BspEI-PstI fragment containing sequences from −92 to +220 (relative to the +1A transcriptional start site) of the fliX promoter was incubated in the presence or absence of CtrA and then digested with DNase I as described in Materials and Methods. The triangle indicates increasing concentrations of His-CtrA (10, 20, and 40 μg). The minus signs indicate that no protein was added, and the asterisk indicates a hypersensitive site. (B) Nucleotide sequence of the fliX-flgI intergenic region showing −10 and −35 regions for fliX and flgI as well as the CtrA binding site and potential cis-acting regulatory elements. Numbers are given relative to the +1A transcriptional start sites. The nucleotide sequence is shown from 5′ to 3′ in the direction of transcription of flgI. Boldface nucleotides denote the consensus CtrA binding site, and ftr denotes the potential binding sites for the transcriptional activator FlbD. Double-underlined sequences at −24 and −12 of the flgI promoter conform to the consensus sequence for ς54-dependent promoters. Overlined is the conserved binding site for integration host factor (IHF). Arrows indicate the transcriptional start sites determined by S1 nuclease analysis. (C) Comparison of the organization of cis-acting elements in the fliX-flgI (26) and fliL-flgF (29, 53) intergenic regions. Arrows denote transcriptional start sites.