Fig. 3 |. Converging RBP–effector interactions regulating (peri-)nuclear RNA processing.
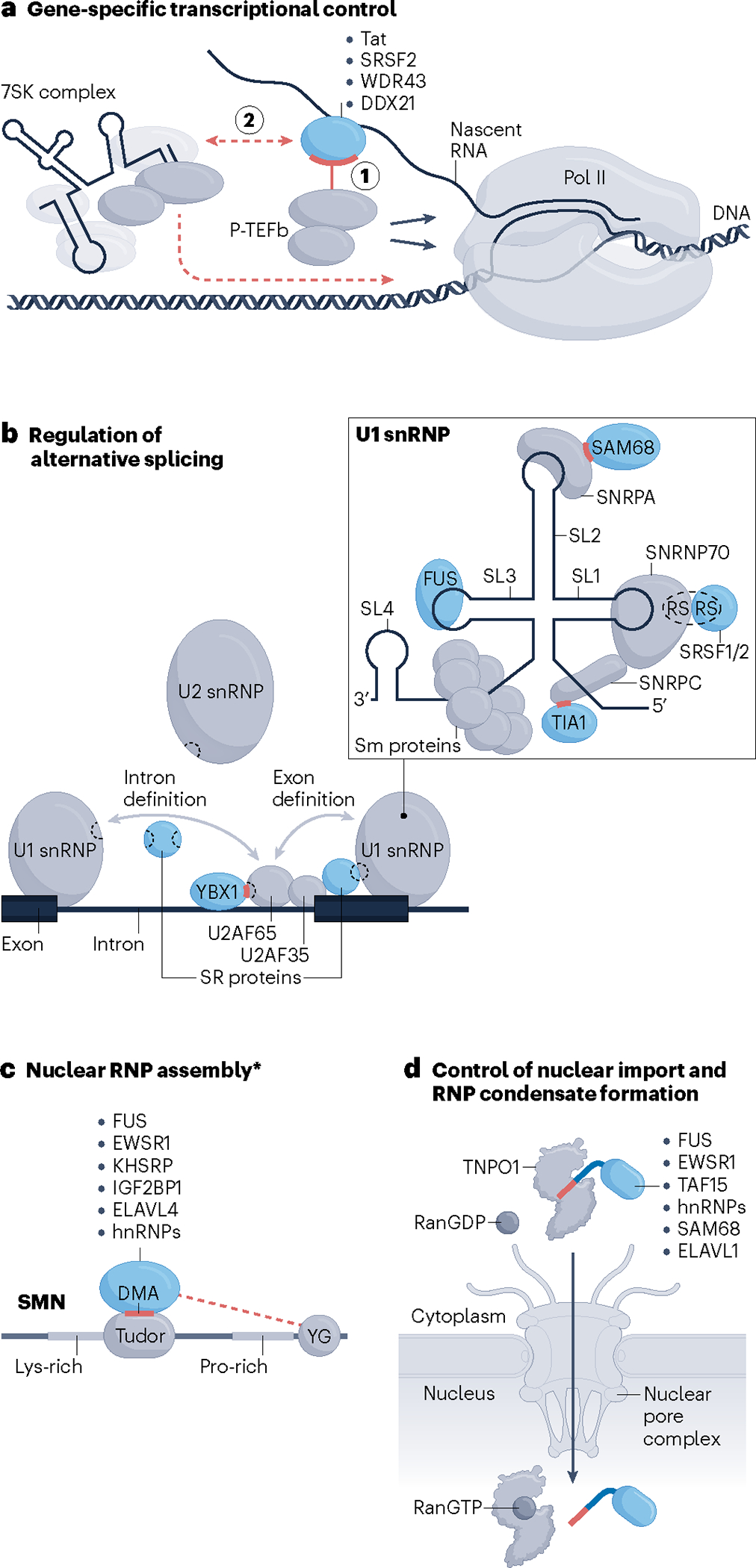
a, RNA-binding protein (RBP)-dependent release of paused RNA polymerase II (Pol II) by positive transcription elongation factor b (P-TEFb). RBPs can stimulate relocation of P-TEFb from a local 7SK complex to the vicinity of Pol II either by recruiting P-TEFb via direct protein–protein interactions (PPIs) while bound to nascent RNA (step 1) or indirectly by associating with or disassociating from the 7SK complex (step 2). b, RBPs directly interact with spliceosomal components, including U1 small nuclear ribonucleoprotein (snRNP), U2 snRNP and U2AF subunits, to promote the early stages of spliceosome assembly. Illustrated is an overview of all interactions (left) along with a zoomed-in view of the U1 snRNP (right). Grey arrows denote intron or exon definition interactions40, several of which are mediated by RS domains (dashed sections of shapes) of SR proteins, such as SRSF1 and SRSF2, and components of the spliceosome. RBPs other than SR proteins, including YBX1, SAM68 and TIA1, use short stretches of their intrinsically disordered regions (IDRs) (red dashes) to contact the indicated spliceosomal proteins. FUS recognizes the stem–loop (SL) region 3 (SL3) of the U1 small nuclear RNA (snRNA). Sm proteins are seven core spliceosomal proteins that make up a stable ring-like structure. c, Dimethylated arginines (DMAs) in the RGG/RG-rich regions of the indicated RBPs are recognized by the aromatic cage within the Tudor domain of survival motor neuron protein (SMN). Dashed red line denotes additional IDR-mediated interactions of some of the listed RBPs with the YG box domain (YG). The asterisk indicates that the same RBP–SMN interactions might also participate in processes other than RNP assembly in the nucleus43. For clarity, RBP-bound RNA is not drawn. d, RBPs that use a proline-tyrosine-rich nuclear localization sequence (PY-NLS) short linear motif (SLiM) (red dash) to interact with transportin 1 (TNPO1) for their nuclear import. The disaggregase activity of TNPO1 is not indicated (Fig. 4e). Drawings of multiple RBPs binding to the same effector molecule in individual panels solely illustrate that different RBPs can bind to a particular effector; they do not imply simultaneous interactions of multiple RBPs with different segments of the same effector or competition between different RBPs for binding to a particular region of an effector molecule.
