ABSTRACT
Porcine reproductive and respiratory syndrome virus (PRRSV) is an important pathogen endangering the swine industry worldwide. Our previous study confirmed that the live attenuated vaccine SD-R strain could provide broad cross-protection against homologous and heterologous NADC30-like viruses. However, it is unclear whether SD-R can provide protection for piglets against HP-PRRSV. In the present study, 4-week-old PRRSV-free piglets were vaccinated intramuscularly with 106.2 TCID50 of SD-R and then challenged intramuscularly (2 mL) and intranasally (2 mL) with HP-PRRSV HuN4 (1 × 105.0 TCID50/mL). The results showed that two of five piglets died in the HuN4 challenge group and five of five immunized piglets survived in the immune challenge group. Five piglets in the HuN4 challenge group had a fever for 6–12 days and showed obvious clinical symptoms, but 3 of 5 piglets had a fever for only 1 day at 5 days post inoculation, and only 1 of the 5 piglets in the immune challenge group had a cough. Compared to challenged control piglets, vaccinated piglets had significantly improved daily weight gain and obviously alleviated pathological lesions. Furthermore, piglets immunized with SD-R had reduced thymus atrophy caused by HP-PRRSV HuN4. These results demonstrate that the lineage 1 PRRSV attenuated vaccine SD-R strain provides protection against lethal HP-PRRSV HuN4 challenge. As such, it is a promising vaccine candidate for use against PRRSV infection.
IMPORTANCE
Both highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) and NADC30-like PRRSV have caused tremendous economic losses to the Chinese pig industry. In this study, a good challenge model was established to evaluate the protection afforded by the candidate SD-R vaccine against infection with a representative HP-PRRSV strain (HuN4). The control piglets in the challenge experiment displayed obvious clinical symptoms of PRRSV infection, with a mortality rate up to 40%. In contrast, all the piglets in the vaccinated challenged group survived, and only some pigs had transient fever. The daily gain of SD-R immunized group piglets was significantly increased, and the pathological changes were significantly reduced. In addition, the viral replication levels in the serum of the immunized group were significantly lower than those of the challenged control group. The live attenuated vaccine SD-R strain can provide protection against HP-PRRSV challenge, indicating that the SD-R strain is a promising vaccine candidate for use in the swine industry.
KEYWORDS: lineage 1, attenuated vaccine, SD-R, HP-PRRSV HuN4, cross-protection efficacy
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS) is one of the most important diseases causing enormous economic losses to the global swine industry. PRRS virus (PRRSV), the aetiological agent of PRRS, is characterized by fast mutation, complex recombination, and large differences in pathogenicity (1). PRRSVs can be divided into two distinct species, Betaarterivirus suid 1 (PRRSV-1) and Betaarterivirus suid 2 (PRRSV-2) (ICTV2021). PRRSV-1 is divided into three or four subtypes {subtype 1 [subtype 1 (Global)], subtype 2 [subtype I (Russia) and subtype II], and subtype 3 [subtype III]} (2 – 6), and PRRSV-2 is divided into nine lineages (lineage 1-lineage 9) (6). Since 1995, subtype I (global) of PRRSV-1 and lineage 1, lineage 3, lineage 5, and lineage 8 of PRRSV-2 have coexisted in China (7). Currently, NADC30-like PRRSV and NADC34-like PRRSV in lineage 1 have become the major endemic strains in China (7 – 12). However, existing PRRSV vaccines provide only limited protection for piglets against lineage 1 PRRSV (13 – 20).
In mainland China, the first PRRSV strain, named CH-1a, belongs to sublineage 8.7 and was first reported in 1996 (21). In 2006, a highly pathogenic PRRSV (HP-PRRSV) containing a discontinuous deletion of 1 + 29 aa in the NSP2 protein caused an outbreak in China that was characterized by high morbidity and mortality (22 – 26). HP-PRRSV causes large numbers of pig deaths and tremendous economic losses in China and some pig-raising countries in Southeast Asia (24, 27, 28). The HP-PRRSV HuN4 strain was isolated and caused 100% morbidity and 40%–100% mortality in piglets (26). Subsequently, HP-PRRSV vaccines (JXA1-P80, HuN4-F112, TJM-92, and GDr180) were developed and applied throughout the country (29 – 31).
PRRSV, characterized by its mutability, produces some novel variants causing outbreaks or recurrent outbreaks in previously vaccinated or unvaccinated swine herds. Heterologous cross-protection by most PRRS vaccines is an obvious deficiency and is also the focus of veterinary researchers. In a previous study, we developed an attenuated lineage 1 PRRSV vaccine (SD-R) that provides safe and effective protection against homologous NADC30-like PRRSV and heterologous NADC30-like PRRSV challenges (32). Furthermore, SD-R could provide effective protection against NADC34-like PRRSV (unpublished data). However, the protective efficacy of SD-R against lethal HP-PRRSV challenge in piglets is unknown.
MATERIALS AND METHODS
Viruses and animals
The candidate live attenuated vaccine SD-R strain was attenuated and stored in our laboratory. The HP-PRRSV HuN4 strain was isolated in 2006 and is maintained in our laboratory. Thirteen 28-day-old confirmed PRRSV-free piglets (antigens of PRRSV, ASFV, CSFV, and PRV were detected using PCR or RT-PCR; antibodies against PRRSV, ASFV, CSFV, and PRV were detected using commercial ELISA kits) were obtained from a PRRS-free farm in Harbin.
Phylogenetic analysis
All sequences were aligned using MAFFT version 7 (33) with default parameters and manually adjusted in MEGA6 (34). Deduced amino acid sequences were aligned with ClustalW with Lasergene software. Phylogenetic trees based on the whole genome were constructed in MEGA 6.0 using the neighbor-joining method with 1,000 bootstrap replicates. The trees were annotated and modified using Evolview (version 2.0) (35).
Evaluation of the immunoprotective effect of SD-R against the HP-PRRSV HuN4 strain
Thirteen PRRSV-free piglets were randomly (the piglets were mixed together before the group assignment) divided into three groups (Groups A, B, and C) (Table 1). Five piglets in group A were used for immunization and inoculation. Five piglets in group B were used for inoculation. The other three piglets in group C were used as negative controls. The piglets in group A were inoculated intramuscularly (2 mL) with 106.2 TCID50/mL SD-R. At 28 days post vaccination (dpv), the piglets in groups A and B were infected intramuscularly (2 mL) and intranasally (2 mL), respectively, with fifth-passage HuN4 (1 × 105.0 TCID50/mL). The animals were maintained in individual biosafety rooms. Clinical signs and rectal temperatures were recorded daily. The body weights of the piglets were measured weekly. Blood samples were periodically collected from individual piglets and tested for viremia. All of the piglets were euthanized at 21 days post inoculation (dpi). Ten tissue samples were obtained from the hearts, livers, spleens, lungs, kidneys, lymph nodes, tonsils, small intestines, bladders, and stomachs for viral detection by TaqMan-based real-time fluorescence quantitative RT-PCR (36).
TABLE 1.
Group information for animal experiments
| Group | Corresponding group | Number of animals | Vaccination | Challenge |
|---|---|---|---|---|
| A | SD-R vaccine-treated and HuN4-challenged group | 5 (091; 092; 093; 094; 035) | 1 × 106.2 TCID50 per pig (SD-R) | 4 × 105.0 TCID50 per pig (HuN4) |
| B | HuN4-challenged group | 5 (062; 063; 064; 065; 066) | DMEM | |
| C | Negative control group | 3 (067; 068; 069) | DMEM |
Serological examination
Serum samples were collected at 0, 7, 14, 21, and 28 dpv and at 3, 5, 7, 10, 14, and 21 dpi. PRRSV-specific antibodies were quantified using a commercial ELISA kit (JNT, Beijing, China) according to the manufacturer’s instructions. The PRRSV-specific antibody titer is reported as the S/P ratio, and the serum samples were considered positive if the S/P ratio was ≥0.4.
To perform serum neutralization (SN) assays, all sera collected at 28 dpv and 21 dpi were heat-inactivated at 56°C for 30 min. Subsequently, serial twofold dilutions of each serum sample were prepared using DMEM as the diluent. Suspensions containing 100 TCID50 of PRRSV per 100 µL were then prepared, and 100 µL of the suspension was added to each serum dilution. The serum-viral mixtures were incubated for 1 h at 37°C in a water bath. Then, the mixtures were dispensed onto MARC-145 cells in 96-well plates. The plates were further incubated at 37°C in a humidified atmosphere with 5% CO2 for 7 days. Duplicate samples were analyzed for CPE (cytopathic effect) daily. The neutralization titer of the serum was calculated using the Reed-Muench method. Three independent tests were performed for each serum sample.
Viremia and viral loads in tissue assessment
To determine the duration of viremia and viral loads in different tissues after treatment with the SD-R vaccine strain, serum samples collected at 0, 7, 14, 21, and 28 dpv and 3, 5, 7, 10, 14, and 21 dpi and 10 tissues of all the piglets were used to detect the RNA copy number of PRRSV by TaqMan-based real-time fluorescence quantitative RT-PCR (36).
Histological examination
At necropsy, the lungs and lymph nodes were harvested and examined for histopathology following hematoxylin and eosin (H&E) staining as previously described (37).
Statistical analysis
Significant differences between two groups were determined using a t test (and nonparametric tests) in GraphPad 5.0 (San Diego, CA, USA). The level of significance was set at P < 0.05.
RESULTS
Genomic characteristics of NADC30-like PRRSV SD-R and HP-PRRSV HuN4
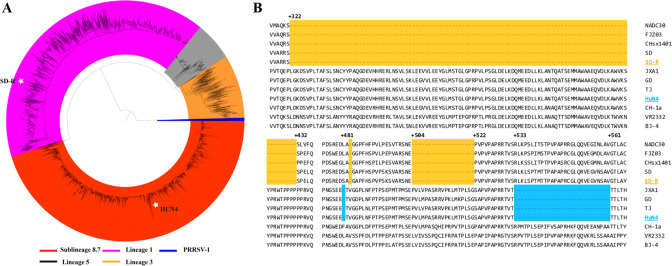
To clarify the relationship between NADC30-like PRRSV SD-R and HP-PRRSV HuN4, the ORF5 sequences of 5,477 PRRSV isolates from China were used to construct a phylogenetic tree. Phylogenetic analysis showed that five subgenotype PRRSVs coexisted in China, including PRRSV-1 subtype I (Global) and PRRSV-2 lineages 1, 3, 5, and 8 (Fig. 1A). SD-R belongs to lineage 1 (1.8), but HuN4 belongs to lineage 8 (8.7) (Fig. 1A). The deduced amino acid alignment of NSP2 showed that SD-R has a 131 aa (111 + 1 + 19) discontinuous deletion and that HuN4 has a 30 aa (1 + 29) discontinuous deletion (Fig. 1B). The genomic nucleotide similarity between SD-R and HuN4 was 83.5%, and the nucleotide similarity and deduced aa similarity among different genes were 75.2%–93.3% and 70.8%–96.7%, respectively (Table 2).
Fig 1.
Phylogenetic analyses and NSP2-deduced amino acid alignment of the SD-R and HuN4 strains. (A) Phylogenetic tree of SD-R and HuN4 based on all the ORF5 sequences of PRRSV. The five lines with different colors represent the five types of PRRSVs. Both SD-R and HuN4 are labeled with white pentagons. (B) The NSP2 deletion pattern in SD-R and HuN4: 131-aa discontinuous deletions are labeled with a light-yellow background; 30-aa discontinuous deletions are labeled with a light blue background.
TABLE 2.
The nucleotide and deduced amino acid similarity between SD-R and HuN4
| Gene | Nucleotide similarity (%) | Deduced amino acid similarity (%) | Gene | Nucleotide similarity (%) | Deduced amino acid similarity (%) |
|---|---|---|---|---|---|
| Whole genome | 83.5 | / | Nsp9 | 86.6 | 96.7 |
| 5′UTR | 89.4 | / | Nsp10 | 84.1 | 95.0 |
| 3′UTR | 93.3 | / | Nsp11 | 89.8 | 95.1 |
| Nsp1α | 87.2 | 94.4 | Nsp12 | 87.1 | 94.8 |
| Nsp1β | 80.0 | 75.2 | ORF2a | 87.8 | 88.3 |
| Nsp2 | 75.2 | 70.8 | ORF2b | 90.5 | 90.5 |
| Nsp3 | 81.3 | 89.1 | ORF3 | 86.7 | 83.5 |
| Nsp4 | 85.0 | 93.1 | ORF4 | 85.8 | 86.6 |
| Nsp5 | 87.5 | 93.5 | ORF5 | 86.5 | 86.9 |
| Nsp6 | 91.7 | 93.8 | ORF5a | 87.0 | 82.6 |
| Nsp7α | 83.0 | 91.3 | ORF6 | 89.4 | 93.1 |
| Nsp7β | 77.3 | 74.5 | ORF7 | 89.2 | 90.3 |
| Nsp8 | 87.4 | 93.3 | N | ||
Clinical reactions after immunization and challenge
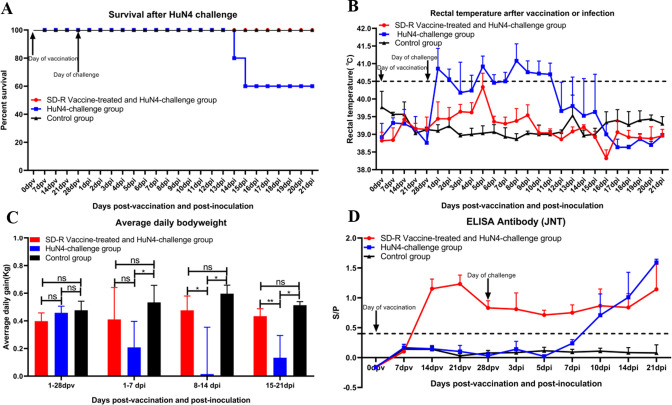
After SD-R immunization, none of the piglets in group A showed any clinical signs of PRRS in contrast to those in group B and group C. After HuN4 challenge, two piglets in group B died at 14 and 15 dpi, but all the piglets in groups A and C survived (Fig. 2A). All the piglets in group B displayed various disease manifestations, including high fever (≥41.0) (Fig. 2B), listlessness, anorexia and emaciation, lying down, and cough. Three piglets had a fever for only 1 day, at 5 dpi (Fig. 2B), and only 1 piglet in group A showed a cough. During the study, the piglets in group C had no clinical signs of disease. After HuN4 challenge, compared to the piglets in groups A and C, the piglets in group B gained less body weight (P < 0.05) from 1 to 7, 8 to 14, and 15 to 21 dpi (Fig. 2C).
Fig 2.
Survival, rectal temperature, average daily bodyweight, and anti-PRRSV antibody levels after SD-R immunization and challenge with the HP-PRRSV HuN4 strain. (A) Survival after HuN4 challenge. (B) Rectal temperatures after SD-R immunization and challenge with the HuN4 strain. (C) Average daily body weight after SD-R immunization and challenge with the HuN4 strain. (D) Anti-PRRSV antibody levels after SD-R immunization and challenge with the HuN4 strain. Tree groups are labeled with red, blue, and black.
Antibody responses in immunized or challenged piglets
The antibody response determined via ELISA showed that all the immunized piglets in group A were seroconverted at 14 dpv (Fig. 2D). A total of 4 of 5 piglets in group B were seroconverted by 10 dpi, and the remaining piglets were seroconverted by 14 dpi (Fig. 2D). No PRRSV-specific antibodies were detected in the control piglets prior to challenge (Fig. 2D). The antibody responses of the piglets in group C were negative throughout the study (Fig. 2D).
Gross pathological and histopathological changes
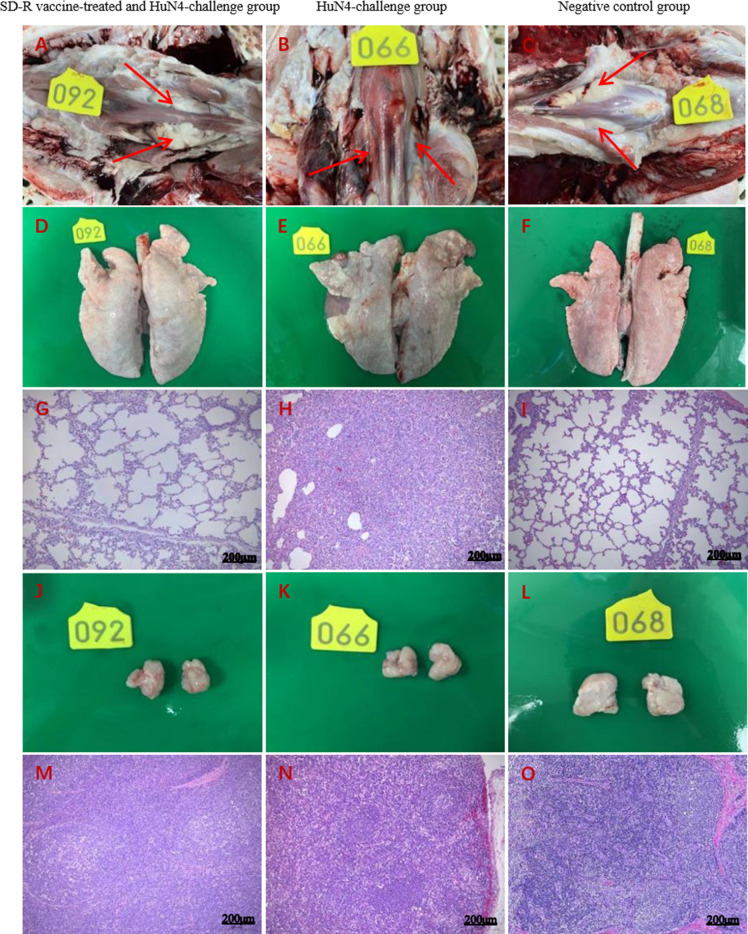
All piglets were euthanized and dissected at 21 dpi (after HuN4 challenge). In contrast with the piglets in groups A (Fig. 3A, D, and J) and C (Fig. 3C, F, and L), the piglets in group B showed lesions typical of PRRS, such as thymus atrophy (Fig. 3B) and consolidation in the lungs (Fig. 3E). Histopathology revealed extensive infiltration of inflammatory cells, loss of alveolar structure over large areas, airway obstruction in the lungs (Fig. 3H), and decreased lymphocyte and medullary bleeding in the lymph nodes (Fig. 3N) in group B compared with those in the negative control groups (Fig. 3I and O). Notably, some piglets in group A showed milder infiltration of inflammatory cells in the lungs (2 of 5) and slightly decreased levels of lymphocytes (3 of 5) (Fig. 3G and M).
Fig 3.
Gross and histological lesions of lungs and lymph nodes after SD-R immunization and challenge with the HP-PRRSV HuN4 strain. In contrast with the piglets in the immunized and challenge group (A, D, and J) and the negative group (C, F, and L), the piglets in the HuN4 challenge group showed lesions typical of PRRS, such as thymus atrophy (B) and consolidation in the lungs (E). Compared with those in the negative control group (I, O), some piglets in the immunized and challenge groups (G, M) showed milder infiltration of inflammatory cells in the lungs and slightly decreased levels of lymphocytes. All the piglets in the HuN4-challenged group showed extensive infiltration of inflammatory cells, loss of alveolar structure over large areas, airway obstruction in the lungs (H), and decreased lymphocyte and medullary bleeding in the lymph nodes (N).
Comparison of viremia and viral tissue distribution between the immunized-challenge group and the challenge group
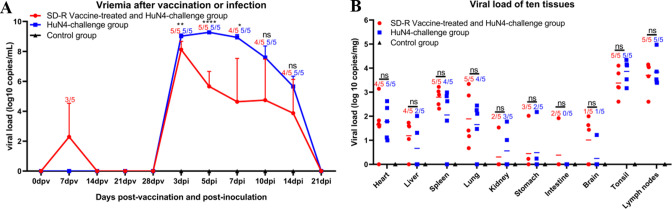
To evaluate the difference in viremia and distribution in 10 tissues among the different groups, serum samples from 0, 7, 14, 21, and 28 dpv; serum samples from 3, 5, 7, 10, 14, and 21 dpi; and 10 organ tissues were evaluated using real-time PCR. The results showed that 3 of the 5 piglets in group A had low levels of viremia at 7 dpv (Fig. 4A). The RNA copy numbers of the serum samples from group B reached their highest levels at 3 dpi and then gradually declined after 10 dpi (Fig. 4A). The viremia levels at 3–7 dpi in group A were significantly lower than those in group B (Fig. 4A). The viral loads of the 10 tissues were not significantly different between group A and group B (Fig. 4B).
Fig 4.
Viremia (A) and viral loads in 10 tissues (B) after SD-R immunization and challenge with the HP-PRRSV HuN4 strain. PRRSV viral RNA in sera and tissues was quantified by qPCR. An asterisk (*) indicates a significant difference between the immunized and challenge groups and the HuN4 challenge group (ns, P > 0.5; *P < 0.05; **P < 0.01; ****P < 0.0001). The numbers represent the number of piglets with viral loads and the number of piglets in the group. The three groups are labeled with red, blue, and black, respectively.
Serum-neutralizing antibody detection
To explore the role of SN Ab in conferring protection, an SN assay was performed using serum samples collected at 28 dpv and 21 dpi from all pigs. CPE was found in all the control wells at 3 days after cell inoculation (100 TCID50/well). In Group A, 5/5 and 1/5 pigs immunized with SD-R (28 dpv) induced homologous SN Ab on days 3 and 7 after cell inoculation, respectively. In addition, all pigs immunized with SD-R (21 dpi) induced homologous SN Ab on days 3 and 7 after cell inoculation (Table 3). In the SN assay against the HuN4 strain, at 28 dpv, neither group A nor group B developed SN Ab against the HuN4 strain either (Table 4). No neutralization titers were detected in group B pigs at 21 dpi. In contrast, serum from group A pigs at 21 dpi could delay the appearance of CPE on the third day of the SN assay, and 3 pigs were able to completely neutralize the HuN4 strain on day 7 (Table 4). No SN Ab was found in the challenge control group and negative control group.
TABLE 3.
Serum neutralization titer of three different group against SD-R strain
| Vaccinated challenged group | Serum neutralization titer | Challenged group | Serum neutralization titer | Negative control group | Serum neutralization titer | |||
|---|---|---|---|---|---|---|---|---|
| 3 days | 7 days | 3 days | 7 days | 3 days | 7 days | |||
| 091 28 dpv | 5.62 | <1:4 | 062 28 dpv | <1:4 | <1:4 | 067 28 dpv | <1:4 | <1:4 |
| 092 28 dpv | 5.62 | <1:4 | 063 28 dpv | <1:4 | <1:4 | 068 28 dpv | <1:4 | <1:4 |
| 093 28 dpv | 6.32 | <1:4 | 064 28 dpv | <1:4 | <1:4 | 069 28 dpv | <1:4 | <1:4 |
| 094 28 dpv | 5.62 | <1:4 | 065 28 dpv | <1:4 | <1:4 | 067 21 dpi | <1:4 | <1:4 |
| 035 28 dpv | 11.34 | 4 | 066 28 dpv | <1:4 | <1:4 | 068 21 dpi | <1:4 | <1:4 |
| 091 21 dpi | 16 | 16 | 062 21 dpi | <1:4 | <1:4 | 069 21 dpi | <1:4 | <1:4 |
| 092 21 dpi | 22.68 | 4 | 063 21 dpi | <1:4 | <1:4 | |||
| 093 21 dpi | 27.12 | 16 | 064 21 dpi | <1:4 | <1:4 | |||
| 094 21 dpi | 32 | 32 | 065 21 dpi | <1:4 | <1:4 | |||
| 035 21 dpi | 40 | 32 | 066 21 dpi | <1:4 | <1:4 | |||
TABLE 4.
Serum neutralization titer of three different group against HuN4 strain
| Vaccinated challenged group | Serum neutralization titer | Challenged group | Serum neutralization titer | Negative control group | Serum neutralization titer | |||
|---|---|---|---|---|---|---|---|---|
| 3 days | 7 days | 3 days | 7 days | 3 days | 7 days | |||
| 091 28 dpv | <1:4 | <1:4 | 062 28 dpv | <1:4 | <1:4 | 067 28 dpv | <1:4 | <1:4 |
| 092 28 dpv | <1:4 | <1:4 | 063 28 dpv | <1:4 | <1:4 | 068 28 dpv | <1:4 | <1:4 |
| 093 28 dpv | <1:4 | <1:4 | 064 28 dpv | <1:4 | <1:4 | 069 28 dpv | <1:4 | <1:4 |
| 094 28 dpv | <1:4 | <1:4 | 065 28 dpv | <1:4 | <1:4 | 067 21 dpi | <1:4 | <1:4 |
| 035 28 dpv | <1:4 | <1:4 | 066 28 dpv | <1:4 | <1:4 | 068 21 dpi | <1:4 | <1:4 |
| 091 21 dpi | 10 | 10 | 062 21 dpi | <1:4 | <1:4 | 069 21 dpi | <1:4 | <1:4 |
| 092 21 dpi | 10 | <1:4 | 063 21 dpi | <1:4 | <1:4 | |||
| 093 21 dpi | 16 | 6.32 | 064 21 dpi | <1:4 | <1:4 | |||
| 094 21 dpi | 5 | <1:4 | 065 21 dpi | <1:4 | <1:4 | |||
| 035 21 dpi | 27.12 | 16 | 066 21 dpi | 8 | <1:4 | |||
DISCUSSION
Both PRRSV-1 and PRRSV-2 are important diseases threatening the normal development of the swine industry (38). Two genotypes (PRRSV-1 and PRRSV-2), including six subgenotypes [subtype I (Global) and lineages 1.5, 1.8, 3.5, 5.1, and 8.7 (C-PRRSV and HP-PRRSV)], coexisted in mainland China (7, 9). Among them, lineages 1.8 (NADC30-like PRRSV) and 8.7 (HP-PRRSV) caused tremendous economic losses to the Chinese pig industry (23, 39). Two types of PRRSV vaccines (lineage 5: RespPRRS MLV/Ingelvac PRRS MLV; lineage 8: CH-1a, CH-1R, JXA1-P80, HuN4-F112, TJM-92, GDr180, PC) have been extensively used in China (15, 29 – 31, 40, 41).
Currently, PRRSV vaccines used in China are all derived from two lineages: lineage 5 (RespPRRS MLV/Ingelvac PRRS MLV) and lineage 8 (CH-1a, CH-1R, JXA1-P80, HuN4-F112, TJM-92, GDr180, PC). The pathogenicity of NADC30-like PRRSV in piglets varies from low to high (31, 39, 42 – 47), but most show moderate pathogenicity (13, 43, 48 – 53). None of the commercial PRRSV vaccines provides complete protection for piglets against NADC30-like PRRSV challenge (13, 14, 16 – 19, 31, 54, 55). The pathogenicity of most HP-PRRSVs is high in piglets (25, 26, 55 – 57). HP-PRRSV vaccines (JXA1-P80, HuN4-F112, TJM-92, and GDr180) could provide complete clinical protection to piglets (15, 29, 31).
Since 2016, NADC30-like PRRSV has become the major endemic strain in China (9, 39). A candidate live attenuated vaccine (SD-R strain) against NADC30-like PRRSV was developed by our laboratory, which has higher safety, provides complete clinical protection for piglets against homologous and heterologous NADC30-like PRRSVs (32) and NADC34-like PRRSV (unpublished), and significantly improves daily weight gain and decreases viremia (32). Thus, SD-R could provide good protection against lineage 1 branch PRRSV. However, it is unknown whether SD-R could provide protection for piglets against a lethal HP-PRRSV challenge. Therefore, in this study, we systematically evaluated the protective efficacy of SD-R against an HP-PRRSV HuN4 strain.
Within 28 days after immunization, none of the immunized piglets showed any clinical signs of PRRS, and all immunized piglets were seroconverted at 14 dpv. There was no significant difference between immunized piglets and negative control piglets in daily weight gain. Only 3 of the 5 immunized piglets had low levels of viremia at 7 dpv, which also further demonstrated that SD-R had a higher safety. After HuN4 challenge, 2 nonimmunized piglets died at 14 and 15 dpi, but all the immunized piglets survived, which means that SD-R could prevent a lethal attack by the HP-PRRSV HuN4 strain. All the nonimmunized piglets displayed various disease manifestations, but only three immunized piglets had a fever for 1 day at 5 dpi, and only 1 immunized piglet showed a cough. Furthermore, piglets immunized with SD-R had improved daily weight gain during the duration of the HuN4 challenge. The above results demonstrated that SD-R could provide at least 80% clinical protection for piglets against HP-PRRSV HuN4 challenge.
Previous research has shown that serum-neutralizing antibodies (SN Abs) play a crucial role in the development of protective immunity against PRRSV (58, 59). Moreover, SN Abs are usually specific for the vaccine strain (homologous), with lower/no titers of cross-neutralizing (heterologous) antibodies (60, 61). In this study, we did not detect neutralizing titers against the HuN4 strain in any 28 dpv serum. Interestingly, we found that the group A pig serum at 21 dpi could delay or completely prevent the HuN4 strain from infecting Marc-145 cells. Thus, although neutralizing titers were not detected at 28 dpv, prophylactic immunization with the SD-R vaccine could help animals generate heterologous neutralizing antibodies in the later stages, reducing the impact of the HuN4 strain on pigs.
Except for death and obvious clinical manifestation, after HP-PRRSV HuN4 challenge, obvious pathological changes, especially thymus atrophy, tissue pathological changes, and high viremia, were observed in the challenged pigs, as observed in previous studies (62 – 67). However, immunized piglets of SD-R could prevent visible pathological damage caused by HuN4, especially preventing thymus atrophy and consolidation in the lungs. Furthermore, piglets immunized with SD-R had significantly alleviated histopathological damage. In addition, piglets immunized with SD-R had significantly reduced viremia, and SD-R helped the pigs produce heterologous neutralizing antibodies in the later stages. At the genus level, SD-R could provide good protection for piglets against lineage 1 and lineage 8 branch PRRSVs, but the protective efficacy against homologous PRRSV was better than that against heterologous PRRSVs. Existing commercial vaccines cannot provide effective protection against other branches of PRRSV strains (13, 19, 32, 41), but SD-R exhibits better cross-protection against different branches of PRRSV strains. Therefore, the NADC30-like PRRSV SD-R strain is a promising vaccine candidate against different PRRSVs. However, the cross-protection mechanism of SD-R needs further study.
Conclusions
The HP-PRRSV HuN4 strain is highly pathogenic in piglets. The lineage 1 PRRSV candidate vaccine strain SD-R produced low-level viremia in some piglets at some time points after immunization. SD-R provided effective protection against lethal attack by the HuN4 strain in piglets, prevented thymus atrophy and consolidation in the lungs, significantly improved daily weight gain and other clinical manifestations, and significantly decreased viremia. SD-R is a promising vaccine candidate against PRRSV.
ACKNOWLEDGMENTS
This study was supported by grants from the Natural Science Foundation of Heilongjiang Province (grant no. YQ2022C042), the National Natural Science Foundation of China (grant nos. 32002315 and 32172890), and the National Center of Technology Innovation for Pigs.
Contributor Information
Zhi-Jun Tian, Email: tianzhijun@caas.cn.
Clinton J. Jones, Oklahoma State University College of Veterinary Medicine, Stillwater, Oklahoma, USA
ETHICS APPROVAL
This study was approved by the Animal Ethics Committee of the School of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences and was performed in accordance with animal ethics guidelines and approved protocols. The Animal Ethics Committee approval number was SYXK (Hei) 2011022.
REFERENCES
- 1. Murtaugh MP, Stadejek T, Abrahante JE, Lam TT, Leung FC. 2010. The ever-expanding diversity of porcine reproductive and respiratory syndrome virus. Virus Res 154:18–30. doi: 10.1016/j.virusres.2010.08.015 [DOI] [PubMed] [Google Scholar]
- 2. Stadejek T, Stankevicius A, Murtaugh MP, Oleksiewicz MB. 2013. Molecular evolution of PRRSV in Europe: current state of play. Vet Microbiol 165:21–28. doi: 10.1016/j.vetmic.2013.02.029 [DOI] [PubMed] [Google Scholar]
- 3. Stadejek T, Oleksiewicz MB, Scherbakov AV, Timina AM, Krabbe JS, Chabros K, Potapchuk D. 2008. Definition of subtypes in the European genotype of porcine reproductive and respiratory syndrome virus: Nucleocapsid characteristics and geographical distribution in Europe. Arch Virol 153:1479–1488. doi: 10.1007/s00705-008-0146-2 [DOI] [PubMed] [Google Scholar]
- 4. Stadejek T, Oleksiewicz MB, Potapchuk D, Podgórska K. 2006. Porcine reproductive and respiratory syndrome virus strains of exceptional diversity in Eastern Europe support the definition of new genetic subtypes. J Gen Virol 87:1835–1841. doi: 10.1099/vir.0.81782-0 [DOI] [PubMed] [Google Scholar]
- 5. Balka G, Podgórska K, Brar MS, Bálint Á, Cadar D, Celer V, Dénes L, Dirbakova Z, Jedryczko A, Márton L, Novosel D, Petrović T, Sirakov I, Szalay D, Toplak I, Leung F-C, Stadejek T. 2018. Genetic diversity of PRRSV 1 in central Eastern Europe in 1994-2014: origin and evolution of the virus in the region. Sci Rep 8:7811. doi: 10.1038/s41598-018-26036-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi M, Lam TT, Hon CC, Hui RK, Faaberg KS, Wennblom T, Murtaugh MP, Stadejek T, Leung FC. 2010. Molecular epidemiology of PRRSV: a Phylogenetic perspective. Virus Res 154:7–17. doi: 10.1016/j.virusres.2010.08.014 [DOI] [PubMed] [Google Scholar]
- 7. Guo Z, Chen XX, Li R, Qiao S, Zhang G. 2018. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol J 15:2. doi: 10.1186/s12985-017-0910-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu H, Li C, Li W, Zhao J, Gong B, Sun Q, Tang YD, Xiang L, Leng C, Peng J, Wang Q, Meng F, Yu Y, An T, Cai X, Tian ZJ, Zhang H. 2022. Novel characteristics of Chinese NADC34‐Like PRRSV during 2020–2021. Transbounding Emerging Dis 69. doi: 10.1111/tbed.14485 [DOI] [PubMed] [Google Scholar]
- 9. Zhang H, Zhang W, Xu H, Song S, Zhao J, Xiang L, Leng C, Li Z, Liu C, Tang Y, Chen J, Peng J, Wang Q, An T, Tong G, Cai X, Tian Z. 2020. The changes of mainly endemic PRRSV in China during 2014-2019. Chin J Prev Vet Med 42:512–516. [Google Scholar]
- 10. Jiang Y, Li G, Yu L, Li L, Zhang Y, Zhou Y, Tong W, Liu C, Gao F, Tong G. 2020. Genetic diversity of porcine reproductive and respiratory syndrome virus (PRRSV). Front. Microbiol 11. doi: 10.3389/fmicb.2020.00618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu F, Yan Y, Shi M, Liu HZ, Zhang HL, Yang YB, Huang XY, Gauger PC, Zhang J, Zhang YH, Tong GZ, Tian ZJ, Chen JJ, Cai XH, Liu D, Li G, An TQ. 2020. Phylogenetics, genomic recombination, and NSP2 polymorphic patterns of porcine reproductive and respiratory syndrome virus in China and the United States in 2014-2018. J Virol 94:e01813-19. doi: 10.1128/JVI.01813-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo Z, Chen XX, Li X, Qiao S, Deng R, Zhang G. 2019. Prevalence and genetic characteristics of porcine reproductive and respiratory syndrome virus in central China during 2016-2017: NADC30-like PRRSVs are predominant. Microb Pathog 135:103657. doi: 10.1016/j.micpath.2019.103657 [DOI] [PubMed] [Google Scholar]
- 13. Zhou L, Yang B, Xu L, Jin H, Ge X, Guo X, Han J, Yang H. 2017. Efficacy evaluation of three modified-live virus vaccines against a strain of porcine reproductive and respiratory syndrome virus NADC30-like. Vet Microbiol 207:108–116. doi: 10.1016/j.vetmic.2017.05.031 [DOI] [PubMed] [Google Scholar]
- 14. Chai W, Liu Z, Sun Z, Su L, Zhang C, Huang L. 2020. Efficacy of two porcine reproductive and respiratory syndrome (PRRS) modified-live virus (MLV) vaccines against heterologous NADC30-like PRRS virus challenge. Vet Microbiol 248:108805. doi: 10.1016/j.vetmic.2020.108805 [DOI] [PubMed] [Google Scholar]
- 15. Zhang H, Xia M, Wang W, Ju D, Cao L, Wu B, Wang X, Wu Y, Song N, Hu J, Tian C, Zhang S, Wu H. 2018. An attenuated highly pathogenic Chinese PRRS viral vaccine confers cross protection to pigs against challenge with the emerging PRRSV NADC30-like strain. Virol Sin 33:153–161. doi: 10.1007/s12250-018-0027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu Y, Zhang Q, Cao Z, Tang YD, Xia D, Wang G, Shan H.. 2021. Recent Advances in Porcine Reproductive and Respiratory Syndrome Virus NADC30-Like Research in China: Molecular Characterization, Pathogenicity, and Control. Front Microbiol 12:791313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei C, Dai A, Fan J, Li Y, Chen A, Zhou X, Luo M, Yang X, Liu J. 2019. Efficacy of type 2 PRRSV vaccine against challenge with the Chinese lineage 1 (NADC30-like) PRRSVs in pigs. Sci Rep 9:10781. doi: 10.1038/s41598-019-47239-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun YF, Zhou L, Bian T, Tian XX, Ren WK, Lu C, Zhang L, Li XL, Cui MS, Yang HC, Yu H. 2018. Efficacy evaluation of two commercial modified-live virus vaccines against a novel recombinant type 2 porcine reproductive and respiratory syndrome virus. Vet Microbiol 216:176–182. doi: 10.1016/j.vetmic.2018.02.016 [DOI] [PubMed] [Google Scholar]
- 19. Bai X, Wang Y, Xu X, Sun Z, Xiao Y, Ji G, Li Y, Tan F, Li X, Tian K. 2016. Commercial vaccines provide limited protection to Nadc30-like PRRSV infection. Vaccine 34:5540–5545. doi: 10.1016/j.vaccine.2016.09.048 [DOI] [PubMed] [Google Scholar]
- 20. Li C, Liu Z, Chen K, Qian J, Hu Y, Fang S, Sun Z, Zhang C, Huang L, Zhang J, Huang N. 2022. Efficacy of the synergy between live-attenuated and inactivated PRRSV vaccines against a NADC30-like strain of porcine reproductive and respiratory syndrome virus in 4-week Piglets. Front Vet Sci 9:812040. doi: 10.3389/fvets.2022.812040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo BQ, Chen ZS, Liu WX, Cui YZ. 1996. Isolation and identification of Procine reproductive and respiratory syndrome virus from aborted fetuses suspected of PRRS. Chin J Anim Poult Infect Dis 87:1–5. [Google Scholar]
- 22. Zhou YJ, Hao XF, Tian ZJ, Tong GZ, Yoo D, An TQ, Zhou T, Li GX, Qiu HJ, Wei TC, Yuan XF. 2008. Highly virulent porcine reproductive and respiratory syndrome virus emerged in China. Transbound Emerg Dis 55:152–164. doi: 10.1111/j.1865-1682.2008.01020.x [DOI] [PubMed] [Google Scholar]
- 23. An TQ, Tian ZJ, Leng CL, Peng JM, Tong GZ. 2011. Highly pathogenic porcine reproductive and respiratory syndrome virus, Asia. Emerg Infect Dis 17:1782–1784. doi: 10.3201/eid1709.110411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng Y, Zhao T, Nguyen T, Inui K, Ma Y, Nguyen TH, Nguyen VC, Liu D, Bui QA, To LT, Wang C, Tian K, Gao GF.. 2008. Porcine respiratory and reproductive syndrome virus variants, Vietnam and China [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang Y, Tian K, Liu W, Fan T, Li F. 2007. Thyroid hormone level change in patients undergoing oral and maxillofacial surgery. Shanghai Kou Qiang Yi Xue 16:611–613. [PubMed] [Google Scholar]
- 26. Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ. 2007. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis 13:1434–1436. doi: 10.3201/eid1309.070399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chaikhumwang P, Tantituvanont A, Tripipat T, Tipsombatboon P, Piriyapongsa J, Nilubol D. 2015. Dynamics and evolution of highly pathogenic porcine reproductive and respiratory syndrome virus following its introduction into a herd concurrently infected with both types 1 and 2. Infect Genet Evol 30:164–174. doi: 10.1016/j.meegid.2014.12.025 [DOI] [PubMed] [Google Scholar]
- 28. Jantafong T, Sangtong P, Saenglub W, Mungkundar C, Romlamduan N, Lekchareonsuk C, Lekcharoensuk P. 2015. Genetic diversity of porcine reproductive and respiratory syndrome virus in Thailand and Southeast Asia from 2008 to 2013. Vet Microbiol 176:229–238. doi: 10.1016/j.vetmic.2015.01.017 [DOI] [PubMed] [Google Scholar]
- 29. Tian ZJ, An TQ, Zhou YJ, Peng JM, Hu SP, Wei TC, Jiang YF, Xiao Y, Tong GZ. 2009. An attenuated live vaccine based on highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) protects piglets against HP-PRRS. Vet Microbiol 138:34–40. doi: 10.1016/j.vetmic.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 30. Liu P, Bai Y, Jiang X, Zhou L, Yuan S, Yao H, Yang H, Sun Z. 2018. High reversion potential of a cell-adapted vaccine candidate against highly pathogenic porcine reproductive and respiratory syndrome. Vet Microbiol 227:133–142. doi: 10.1016/j.vetmic.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 31. Liu C, Zhang W, Gong W, Zhang D, She R, Xu B, Ning Y. 2015. Comparative respiratory pathogenicity and dynamic tissue distribution of Chinese highly pathogenic porcine reproductive and respiratory syndrome virus and its attenuated strain in piglets. J Comp Pathol 153:38–49. doi: 10.1016/j.jcpa.2015.04.002 [DOI] [PubMed] [Google Scholar]
- 32. Zhang H, Xiang L, Xu H, Li C, Tang YD, Gong B, Zhang W, Zhao J, Song S, Peng J, Wang Q, An T, Cai X, Tian ZJ. 2022. Lineage 1 porcine reproductive and respiratory syndrome virus attenuated live vaccine provides broad cross-protection against homologous and heterologous NADC30-like virus challenge in piglets. Vaccines (Basel) 10:752. doi: 10.3390/vaccines10050752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He Z, Zhang H, Gao S, Lercher MJ, Chen W-H, Hu S. 2016. Evolview V2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res 44:W236–41. doi: 10.1093/nar/gkw370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei T, Tian Z, An T, Zhou Y, Xiao Y, Jiang Y, Hao X, Zhang S, Peng J, Qiu H, Tong G. 2008. Development and application of Taq man-MGB fluorescence quantitative RT-PCR assay for detection of porcine reproductive and respiratory syndrome virus.Chin J Prev Vet Med 30:944–991. [Google Scholar]
- 37. Zhang H, Leng C, Tian Z, Liu C, Chen J, Bai Y, Li Z, Xiang L, Zhai H, Wang Q, Peng J, An T, Kan Y, Yao L, Yang X, Cai X, Tong G. 2018. Complete genomic characteristics and pathogenic analysis of the newly emerged classical swine fever virus in China. BMC Vet Res 14:204. doi: 10.1186/s12917-018-1504-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. VanderWaal K, Deen J. 2018. Global trends in infectious diseases of swine. Proc Natl Acad Sci U S A 115:11495–11500. doi: 10.1073/pnas.1806068115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang H, Leng C, Ding Y, Zhai H, Li Z, Xiang L, Zhang W, Liu C, Li M, Chen J, Bai Y, Kan Y, Yao L, Peng J, Wang Q, Tang YD, An T, Cai X, Tian Z, Tong G. 2019. Characterization of newly emerged NADC30-like strains of porcine reproductive and respiratory syndrome virus in China. Arch Virol 164:401–411. doi: 10.1007/s00705-018-4080-7 [DOI] [PubMed] [Google Scholar]
- 40. Chen N, Li X, Xiao Y, Li S, Zhu J. 2021. Characterization of four types of MLV-derived porcine reproductive and respiratory syndrome viruses isolated in unvaccinated pigs from 2016 to 2020. Res Vet Sci 134:102–111. doi: 10.1016/j.rvsc.2020.12.007 [DOI] [PubMed] [Google Scholar]
- 41. Zhou L, Ge X, Yang H. 2021. “Porcine reproductive and respiratory syndrome modified live virus vaccine: a "leaky" vaccine with debatable efficacy and safety”. Vaccines 9:362. doi: 10.3390/vaccines9040362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han G, Lei K, Xu H, He F. 2020. Genetic characterization of a novel recombinant PRRSV2 from lineage 8, 1 and 3 in China with significant variation in replication efficiency and cytopathic effects. Transbound Emerg Dis 67:1574–1584. doi: 10.1111/tbed.13491 [DOI] [PubMed] [Google Scholar]
- 43. Brockmeier SL, Loving CL, Vorwald AC, Kehrli ME, Baker RB, Nicholson TL, Lager KM, Miller LC, Faaberg KS. 2012. Genomic sequence and virulence comparison of four type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res 169:212–221. doi: 10.1016/j.virusres.2012.07.030 [DOI] [PubMed] [Google Scholar]
- 44. Chen N, Ye M, Li S, Huang Y, Zhou R, Yu X, Tian K, Zhu J. 2018. Emergence of a novel highly pathogenic recombinant virus from three lineages of porcine reproductive and respiratory syndrome virus 2 in China 2017. Transbound Emerg Dis 65:1775–1785. doi: 10.1111/tbed.12952 [DOI] [PubMed] [Google Scholar]
- 45. Zhao K, Ye C, Chang XB, Jiang CG, Wang SJ, Cai XH, Tong GZ, Tian ZJ, Shi M, An TQ. 2015. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J Virol 89:10712–10716. doi: 10.1128/JVI.01446-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu JK, Zhou X, Zhai JQ, Li B, Wei CH, Dai AL, Yang XY, Luo ML. 2017. Emergence of a novel highly pathogenic porcine reproductive and respiratory syndrome virus in China. Transbound Emerg Dis 64:2059–2074. doi: 10.1111/tbed.12617 [DOI] [PubMed] [Google Scholar]
- 47. Liu J, Zhou X, Zhai J, Wei C, Dai A, Yang X, Luo M. 2017. Recombination in JXA1-R vaccine and NADC30-like strain of porcine reproductive and respiratory syndrome viruses. Vet Microbiol 204:110–120. doi: 10.1016/j.vetmic.2017.04.017 [DOI] [PubMed] [Google Scholar]
- 48. Sun Z, Wang J, Bai X, Ji G, Yan H, Li Y, Wang Y, Tan F, Xiao Y, Li X, Tian K. 2016. Pathogenicity comparison between highly pathogenic and NADC30-like porcine reproductive and respiratory syndrome virus. Arch Virol 161:2257–2261. doi: 10.1007/s00705-016-2883-y [DOI] [PubMed] [Google Scholar]
- 49. Wang HM, Liu YG, Tang YD, Liu TX, Zheng LL, Wang TY, Liu SG, Wang G, Cai XH. 2018. A natural recombinant PRRSV between HP-PRRSV JXA1-like and NADC30-like strains. Transbound Emerg Dis 65:1078–1086. doi: 10.1111/tbed.12852 [DOI] [PubMed] [Google Scholar]
- 50. Bian T, Sun Y, Hao M, Zhou L, Ge X, Guo X, Han J, Yang H. 2017. A recombinant type 2 porcine reproductive and respiratory syndrome virus between NADC30-like and a MLV-like: genetic characterization and pathogenicity for piglets. Infect Genet Evol 54:279–286. doi: 10.1016/j.meegid.2017.07.016 [DOI] [PubMed] [Google Scholar]
- 51. Zhou L, Kang R, Yu J, Xie B, Chen C, Li X, Xie J, Ye Y, Xiao L, Zhang J, Yang X, Wang H. 2018. Genetic characterization and pathogenicity of a novel recombined porcine reproductive and respiratory syndrome virus 2 among Nadc30-like, Jxa1-like, and Mlv-like strains. Viruses 10:551. doi: 10.3390/v10100551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sui X, Guo X, Jia H, Wang X, Lin W, Li M, Gao X, Wu J, Jiang Y, Willems L, Zhu H, Xin T, Hou S. 2018. Genomic sequence and virulence of a novel NADC30-like porcine reproductive and respiratory syndrome virus isolate from the hebei province of China. Microb Pathog 125:349–360. doi: 10.1016/j.micpath.2018.08.048 [DOI] [PubMed] [Google Scholar]
- 53. Zhou L, Kang R, Zhang Y, Yu J, Xie B, Chen C, Li X, Chen B, Liang L, Zhu J, Tian Y, Yang X, Wang H. 2019. Emergence of two novel recombinant porcine reproductive and respiratory syndrome viruses 2 (lineage 3) in southwestern China. Vet Microbiol 232:30–41. doi: 10.1016/j.vetmic.2019.01.026 [DOI] [PubMed] [Google Scholar]
- 54. Renukaradhya GJ, Meng XJ, Calvert JG, Roof M, Lager KM. 2015. Live porcine reproductive and respiratory syndrome virus vaccines: current status and future direction. Vaccine 33:4069–4080. doi: 10.1016/j.vaccine.2015.06.092 [DOI] [PubMed] [Google Scholar]
- 55. Leng X, Li Z, Xia M, Li X, Wang F, Wang W, Zhang X, Wu H. 2012. Mutations in the genome of the highly pathogenic porcine reproductive and respiratory syndrome virus potentially related to attenuation. Vet Microbiol 157:50–60. doi: 10.1016/j.vetmic.2011.12.012 [DOI] [PubMed] [Google Scholar]
- 56. Tornimbene B, Frossard JP, Chhim V, Sorn S, Guitian J, Drew TW. 2015. Emergence of highly pathogenic porcine reproductive and respiratory syndrome (HP-PRRS) in medium-scale swine farms in southeastern Cambodia. Prev Vet Med 118:93–103. doi: 10.1016/j.prevetmed.2014.08.009 [DOI] [PubMed] [Google Scholar]
- 57. Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P. 2007. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the mid-Eastern region of China. Vet J 174:577–584. doi: 10.1016/j.tvjl.2007.07.032 [DOI] [PubMed] [Google Scholar]
- 58. Osorio FA, Galeota JA, Nelson E, Brodersen B, Doster A, Wills R, Zuckermann F, Laegreid WW. 2002. Passive transfer of virus-specific antibodies confers protection against reproductive failure induced by a virulent strain of porcine reproductive and respiratory syndrome virus and establishes sterilizing immunity. Virology 302:9–20. doi: 10.1006/viro.2002.1612 [DOI] [PubMed] [Google Scholar]
- 59. Lopez OJ, Oliveira MF, Garcia EA, Kwon BJ, Doster A, Osorio FA. 2007. Protection against porcine reproductive and respiratory syndrome virus (PRRSV) infection through passive transfer of PRRSV-neutralizing antibodies is dose dependent. Clin Vaccine Immunol 14:269–275. doi: 10.1128/CVI.00304-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou L, Ni YY, Piñeyro P, Sanford BJ, Cossaboom CM, Dryman BA, Huang YW, Cao DJ, Meng XJ. 2012. DNA shuffling of the GP3 genes of porcine reproductive and respiratory syndrome virus (PRRSV) produces a Chimeric virus with an improved cross-neutralizing ability against a heterologous PRRSV strain. Virology 434:96–109. doi: 10.1016/j.virol.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 61. Vu HL, Kwon B, Yoon KJ, Laegreid WW, Pattnaik AK, Osorio FA. 2011. Immune evasion of porcine reproductive and respiratory syndrome virus through lygcan shielding involves both glycoprotein 5 as well as glycoprotein 3. J Virol 85:5555–5564. doi: 10.1128/JVI.00189-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang G, Song T, Yu Y, Liu Y, Shi W, Wang S, Rong F, Dong J, Liu H, Cai X, Zhou EM. 2011. Immune responses in piglets infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol 142:170–178. doi: 10.1016/j.vetimm.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 63. He Y, Wang G, Liu Y, Shi W, Han Z, Wu J, Jiang C, Wang S, Hu S, Wen H, Dong J, Liu H, Cai X. 2012. Characterization of thymus atrophy in piglets infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Vet Microbiol 160:455–462. doi: 10.1016/j.vetmic.2012.05.040 [DOI] [PubMed] [Google Scholar]
- 64. Wang G, Yu Y, Tu Y, Li Y, Tong J, Zhang C, Liu Y, Li A, Jiang C, Wang S, Tao Y, Cai X. 2015. Characterizing the thymic lesions in piglets infected with attenuated strains of highly pathogenic porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol 168:258–261. doi: 10.1016/j.vetimm.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 65. Li Y, Wang G, Liu Y, Tu Y, He Y, Wang Z, Han Z, Li L, Li A, Tao Y, Cai X. 2014. Identification of apoptotic cells in the thymus of Piglets infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Virus Research 189:29–33. doi: 10.1016/j.virusres.2014.04.011 [DOI] [PubMed] [Google Scholar]
- 66. Wang G, Yu Y, Tu Y, Tong J, Liu Y, Zhang C, Chang Y, Wang S, Jiang C, Zhou EM, Cai X, Leung FC. 2015. Highly pathogenic porcine reproductive and respiratory syndrome virus infection induced apoptosis and autophagy in thymi of infected piglets. PLoS ONE 10:e0128292. doi: 10.1371/journal.pone.0128292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang G, Yu Y, Cai X, Zhou EM, Zimmerman JJ. 2020. Effects of PRRSV infection on the porcine thymus. Trends Microbiol 28:212–223. doi: 10.1016/j.tim.2019.10.009 [DOI] [PubMed] [Google Scholar]