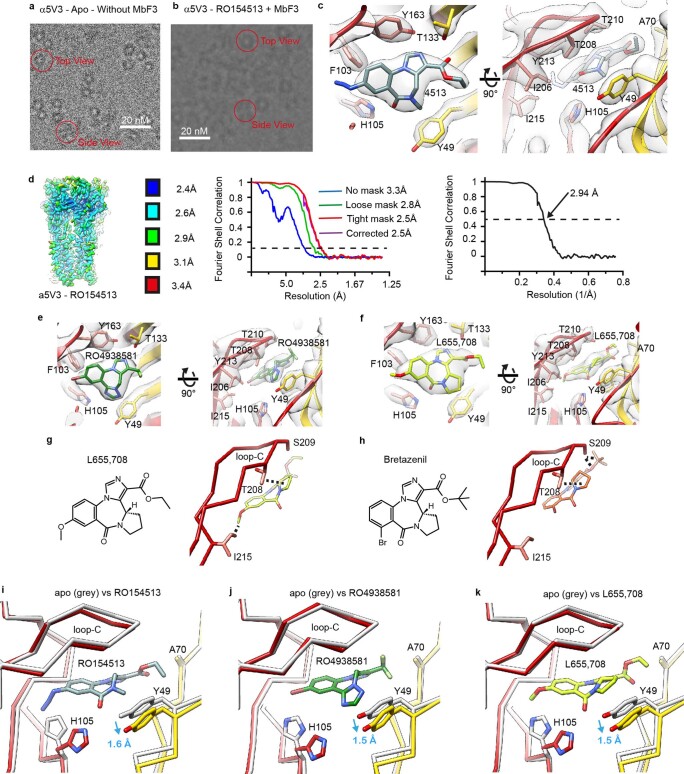
Extended Data Fig. 8. Type II BZD binding modes and impacts.
a, Micrograph showing particle distribution for α5V3-apo, being mostly views, versus, b, for α5V3-RO154513 bound by megabody MbF3, which binds nanodisc MSP2N2, giving mostly side views. Each sample set-up once for data collection. c, Cryo-EM map electron density and fitted protein model of α5V3-RO154513. Shown from two viewing angles. Protein and ligand contour level are the same. d, α5V3-RO154513 map coloured by local resolution (see Methods). Fourier shell correlation (FSC) (upper right panel) and map-model FSC (lower right panel) plots are also shown. Relevant statistics are presented in Data Table 2. e, f, Cryo-EM map electron density and fitted protein models of α5V3-RO154513 and α5V3-L655,708 respectively. Shown from two viewing angles. Protein and ligand contour level are the same. g,h, Structural formula of L655,708 and bretazenil respectively along with Cα stick representation of α5V3 loop-C showing the unique α5 residues T208 and I215 and putative vdW interactions (dashed lines). For bretazenil, h, it does not interact with the unique α5 I215 methyl, and has additional putative interactions with S209 (versus L655,708) due to its larger trimethyl head that could compensate for loss of the unique α5 T208 methyl, to explain why bretazenil is non-selective. i-k, Structural model overlays of α5V3-apo (grey) versus, i, α5V3-RO154513, j, α5V3-RO4938581, and k, α5V3-L655,708, showing that Y49 moves to accommodate binding. An energetic requirement for this structural motion could contribute to RO154513, RO4938581 and L655,708 having 360-fold, 100-fold and 340-fold lower binding affinity respectively for α5V3 versus α5β3γ2 (Extended Data Fig. 4a). For reference, equivalent complementary face residue numbering of α5V3 Y49, A70, T133, in wild type γ2 is Y58, A79, T142 respectively.