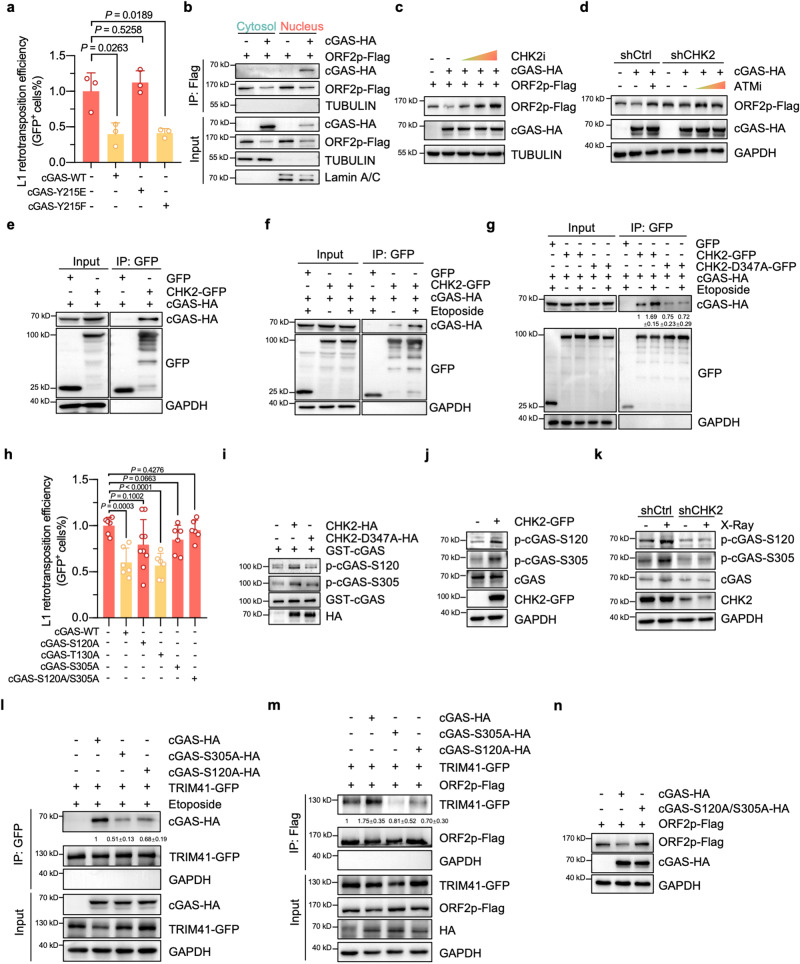
Fig. 3. CHK2 involves in nuclear cGAS-mediated ORF2p degradation and phosphorylates cGASS120/S305 to potentiate its repressive effect on L1 retrotransposition upon DNA damage.
a Analysis of L1 retrotransposition efficiency in HeLa cells expressing cGAS WT or mutants. n = 3 independent experiments. b cGAS interacts with ORF2p in the nucleus. c The effect of a CHK2 inhibitor on cGAS-mediated downregulation at the ORF2p protein level. Cells were treated with the CHK2 inhibitor 6 h post-transfection. d The effect of an ATM inhibitor on cGAS-mediated repression of ORF2p protein levels in CHK2-knockdown HeLa cells. Cells were treated with ATM inhibitor 6 h post-transfection. e Co-IP analysis of the interaction between cGAS and CHK2 in HEK293T cells. f Co-IP analysis showing the interaction between cGAS and CHK2 in HEK293T cells when the DNA was damaged. Cells were treated with 100 µg/mL etoposide for 4 h. g Comparison of the interaction between cGAS and CHK2-WT or CHK2-D347A. h Analysis of L1 retrotransposition efficiency in cells overexpressing WT cGAS and the S120A, T130A and S305A mutants. n = 9 independent experiments for the S120A group, and n = 6 independent experiments for the other groups. i In vitro analysis of CHK2-mediated phosphorylation of cGAS at the S120 or S305 residue. j The effect of CHK2 overexpression on cGAS phosphorylation levels at the S120 or S305 residue. k The effect of CHK2 depletion on cGAS phosphorylation levels at the S120 or S305 residues after DNA was damaged. Cells were treated with 10 Gy X-ray and lysed 2 h post irradiation. l Comparison of the interaction between TRIM41 and cGAS or its mutants after DNA was damaged. Cells were treated with 100 μg/mL etoposide for 4 h. m The effect of overexpressing cGAS or its mutants on the TRIM41-ORF2p interaction in HEK293T cells. n The effect of overexpressing cGAS or its mutants on ORF2p protein levels. Data are presented as mean values ± s.d. Student’s t test was performed for (a) and (h). All the Inputs and IPs were from the same experiments. All statistical test used were two-sided. Experiments were repeated three times independently with similar results.