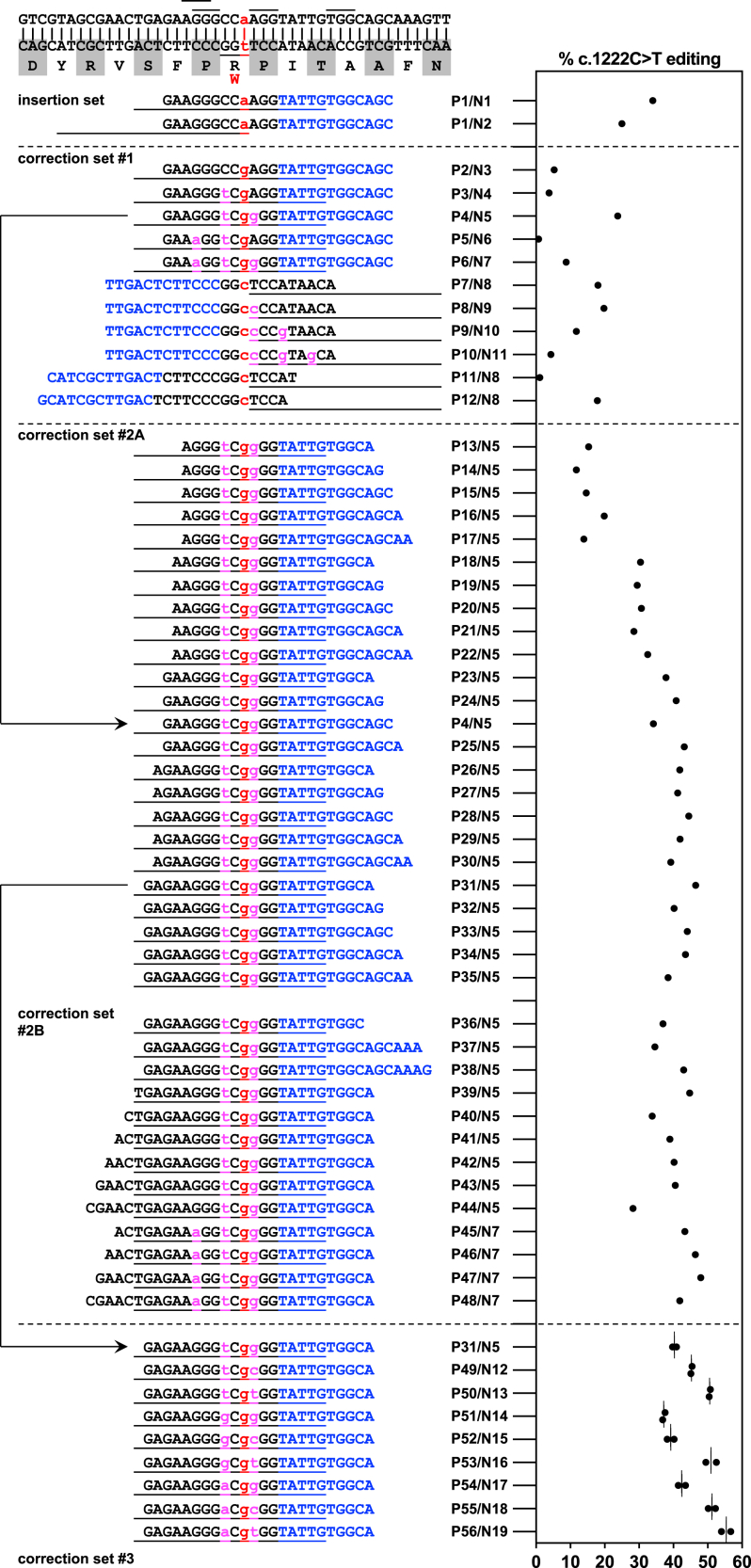
Figure 4.
Prime editing to correct the PAH c.1222C>T variant in human hepatocytes in vitro
Efficiencies of PAH c.1222C>T editing by various pegRNA/ngRNA combinations (Px/Nx, refer to Tables S1 and S2 for sequences); the first two combinations in the table/graph were for insertion of the c.1222C>T variant in wild-type HuH-7 cells (used to generate the PAH c.1222C>T homozygous HuH-7 cell line in Figure 3), and the remainder for correction of the variant in homozygous HuH-7 cells. For each letter sequence, blue indicates the PBS of the pegRNA, and black indicates the RTT of the pegRNA. Red lowercase letters indicate edits at the site of the variant; magenta lowercase letters indicate additional synonymous edits. The short lines at the top of the table indicate NGG PAM sequences used for pegRNAs and/or ngRNAs. Each long line immediately below a pegRNA PBS/RTT sequence in the table indicates the span of the protospacer sequence of the ngRNA used with that pegRNA. n = 1 biological replicate for each pegRNA/ngRNA combination except for the last set, for which n = 2 biological replicates. Lines in the graph = mean values.