Figure 1.
Intron Jaccard index improves splicing outlier calling
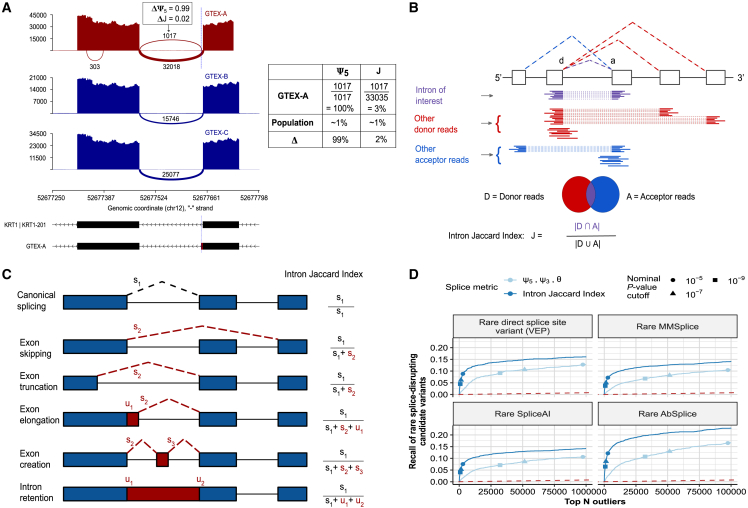
(A) Sashimi plot of three GTEx skin not-sun-exposed RNA-seq samples showing exons 4 and 5 of KRT1. A splicing outlier was detected in the top sample with the ψ5 metric of FRASER (red). The position of the donor site of the outlier intron is indicated with a blue dashed line. Although this intron is not expressed in most other samples (dark blue) and is therefore detected with a high Δψ5 value as shown in the table on the right, its functional impact is probably minor because the canonical intron remains largely dominant.
(B) Schematic definition of the intron Jaccard index for an intron of interest (purple) defined by a donor site d and acceptor site a. The set of donor-associated reads D (red) and acceptor-associated reads A (blue) are highlighted.
(C) Representation of different types of aberrant splicing events that can be captured with the intron Jaccard index. The right column contains the formulae to compute the intron Jaccard index of the canonical intron (black dotted line) from the split (s) and non-split (u) reads of the involved introns in each scenario.
(D) Recall of rare splice-disrupting candidate variants as defined by VEP (canonical splice-site variants, n = 1,544), MMSplice (n = 3,395), SpliceAI (n = 2,971), and AbSplice (n = 2,265) versus the rank of nominal p values from FRASER (light blue) and from an adaptation of FRASER using the intron Jaccard index (dark blue) on the GTEx skin not-sun-exposed dataset (n = 582). Different nominal p value cutoffs are indicated with shapes.