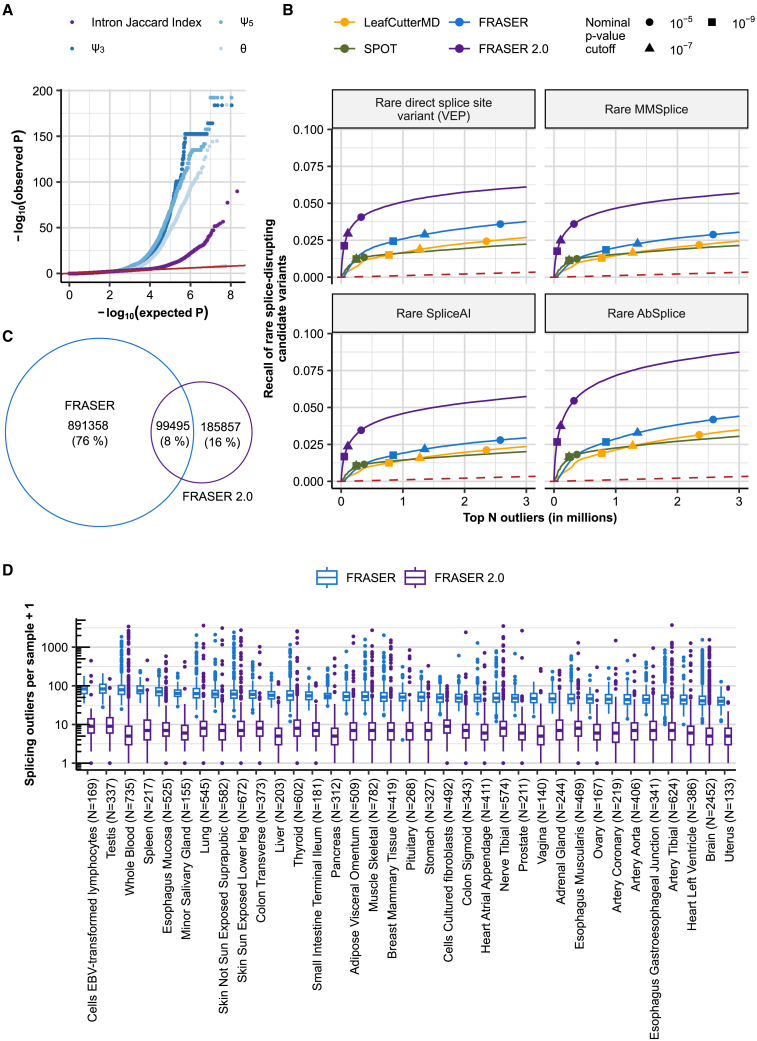
Figure 2.
FRASER 2.0 increases recall of rare splice-disrupting candidate variants on GTEx
(A) Quantile-quantile plots of the p values for the different splice metrics from FRASER (ψ3, ψ5, θ, shown in shades of blue) and the intron Jaccard index from FRASER 2.0 (purple) on the GTEx skin not-sun-exposed dataset. The red line depicts the diagonal, and the gray ribbon around it depicts the 95% confidence interval.
(B) Recall of rare splice-disrupting candidate variants as defined by the variant annotation tools VEP, MMSplice, SpliceAI, and AbSplice (facets) versus the rank of nominal p values combined across GTEx tissues for FRASER (blue), FRASER 2.0 (purple), LeafCutterMD (yellow), and SPOT (green). Nominal p value cutoffs are indicated with shapes.
(C) Venn diagram of the overlap of gene-level splicing outliers found with FRASER (blue) and FRASER 2.0 (purple).
(D) Box plots of the number of splicing outliers (gene level) per sample (y axis) called by FRASER (blue) and FRASER 2.0 (purple) for each GTEx tissue (x axis). All brain tissues have been combined for readability.