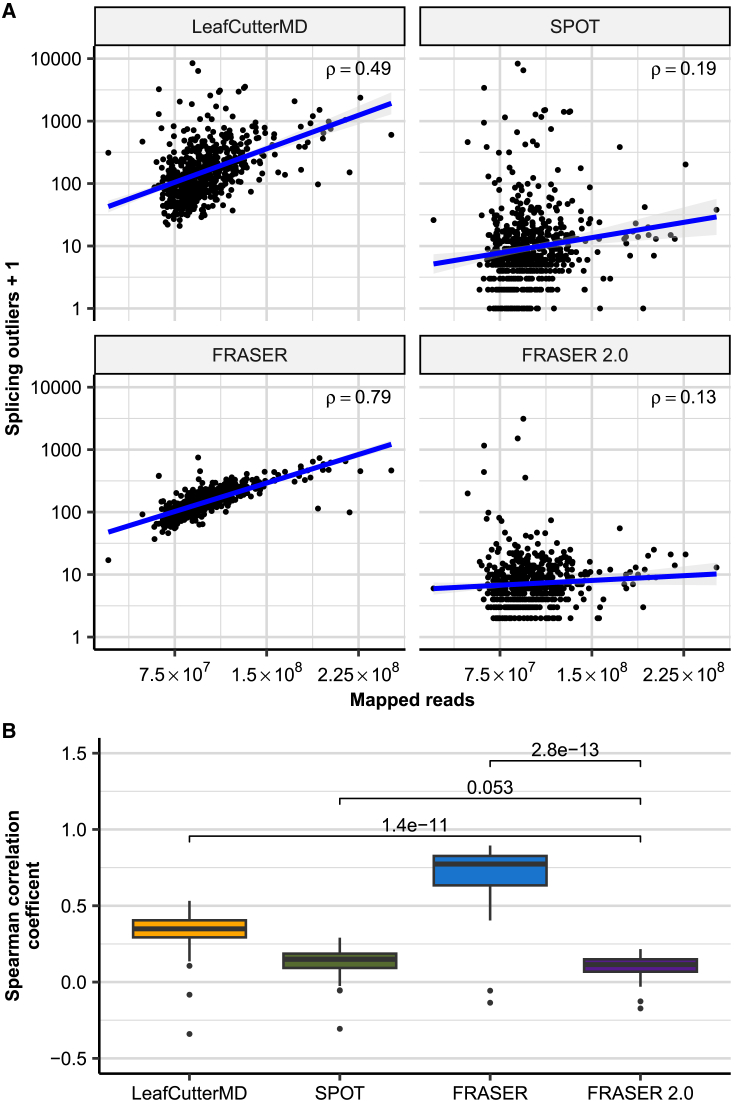
Figure 3.
FRASER 2.0 is less sensitive to sequencing depth than previous methods
(A) Scatterplot of the number of splicing outliers at the gene level against the total mapped reads per sample on the GTEx skin not-sun-exposed dataset for LeafCutterMD, SPOT, FRASER, and FRASER 2.0 (facets). Spearman correlation coefficients (rho) are shown. All are significant (Spearman test, p < 3 × 10−3).
(B) Box plots of the Spearman correlation coefficients (y axis) between the mapped reads and the number of gene-level splicing outliers called by LeafCutterMD, SPOT, FRASER, and FRASER 2.0 (x axis) for each GTEx tissue (n = 48). p values of Wilcoxon tests are shown above brackets.