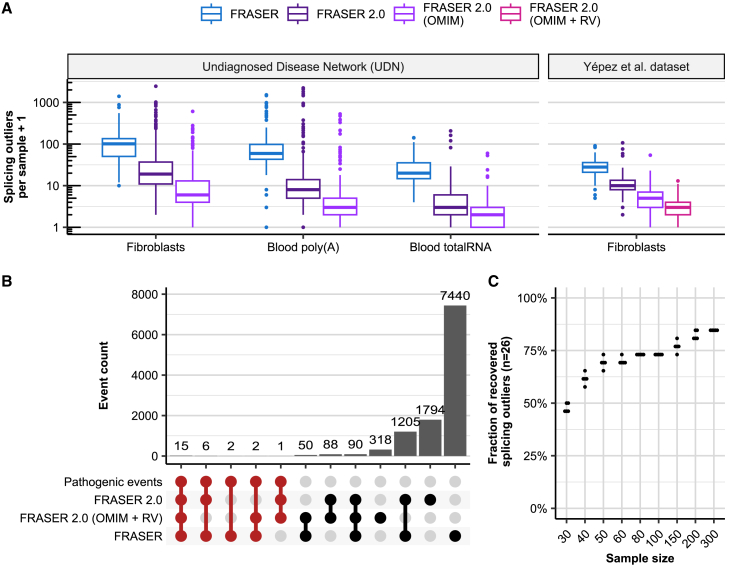
Figure 4.
Application of FRASER 2.0 to rare-disease cohorts
(A) Distribution of the splicing outliers per sample at the gene level on the UDN (n = 391, n = 252, n = 104 for fibroblasts, blood poly(A), and blood total RNA, respectively) and the Yépez et al. dataset (n = 303) for FRASER (blue) and FRASER 2.0 applied to three gene sets considered for FDR correction: expressed genes (dark purple), expressed OMIM genes (light purple), and expressed OMIM genes with a rare variant (violet red; see material and methods).
(B) Number of events (bars) in all non-empty intersections (linked dots) between four splicing outlier sets from the Yépez et al. dataset: (1) the 26 originally reported pathogenic events, (2) the transcriptome-wide significant FRASER 2.0 calls, (3) the significant FRASER 2.0 calls when only OMIM genes with a rare variant are considered, and (4) the transcriptome-wide significant FRASER calls. Intersections with the set of pathogenic events are highlighted in red.
(C) Fraction of recovered pathogenic splicing outliers from the Yépez et al. dataset (y axis, total n = 26) when subsampling to different sample sizes (x axis) was performed. Each sample size was randomly sampled five times. RV: rare variant.