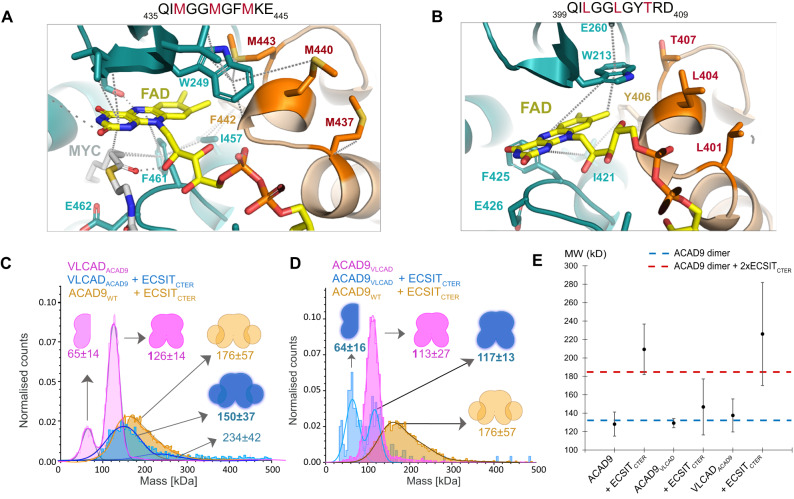
Fig. 5. Identification of the features that distinguishes ACAD9 from VLCAD.
A Three Methionine residues (M437, M440, M443) are situated at the top of the VLCAD FAD site and contribute to the stability of the FAD molecule by M440 and M443 forming sulfur-π interactions with the isoalloxazine moiety and W209. B Unlike VLCAD, ACAD9 has a Threonine and two Leucine residues (in red) and is unable to provide stability to the FAD molecule. C Mass photometry analysis shows that when VLCAD is mutated to mimic ACAD9 (VLCADACAD9, pink), there is a reduction in stability compared to VLCADWT (Fig. 4C). When reconstituted with ECSITCTER, there is a shift in the MW, indicating the formation of a VLCADACAD9-ECSITCTER complex (in blue). D The reverse mutation of ACAD9 to mimic VLCAD (ACAD9VLCAD) produces a more stable dimer (pink). Upon reconstitution of ECSITCTER, we see an appearance of a monomeric ACAD9VLCAD (blue) indicating the destabilisation of the ACAD9 dimer and no significant shift to a higher MW, demonstrating a stark reduction in the ability of ACAD9VLCAD to form a complex with ECSITCTER in comparison to WT (orange). Experiments were repeated thrice with similar results. E DLS measurements of ACAD9WT, ACAD9VLCAD and VLCADACAD9 show similar behaviour. However, in the case of ACAD9VLCAD, there is a significant reduction in complex formation in comparison to ACADWT-ECSITCTER, implying that this mutation does indeed evoke VLCAD-like tendencies. The reverse is also true in the case of VLCADACAD9, a large average MW is seen, indicating the formation of a VLCADACAD9-ECSITCTER complex. This indicates that VLCADACAD9 behaves less like VLCADWT and more like ACAD9WT through the mutation of these key residues. Blue and red dashed lines indicate the expected MW for ACAD9 homodimer and ACAD9WT-ECSITCTER complex, respectively. Data are presented as mean values ± SD of at least n = 3 biological replicas. Source data are provided as a Source Data file.