Summary
In this perspective we discuss the current lack of genetic and environmental diversity in functional genomics datasets. There is a well-described Eurocentric bias in genetic and functional genomic research that has a clear impact on the benefit this research can bring to underrepresented populations. Current research focused on genetic variant-to-function experiments aims to identify molecular QTLs, but the lack of data from genetically diverse individuals has limited analyses to mostly populations of European ancestry. Although some efforts have been established to increase diversity in functional genomic studies, much remains to be done to consistently generate data for underrepresented populations from now on. We discuss the major barriers for this continuity and suggest actionable insights, aiming to empower research and researchers from underserved populations.
There is a dearth of genetic and environmental diversity in functional genomics datasets. Research focused on the variant-to-function gap aims to identify molecular QTLs but lacks data from non-European-ancestry populations. We discuss the major barriers and pose actionable suggestions, which aim to empower research and researchers from underserved populations.
Background
After decades of human genetics research focusing largely on populations of European ancestry, the field has recently begun emphasizing and investing in research that encompasses a broader spectrum of environmental and human ancestral diversity, with the twin aims of diversifying existing human genetics resources and developing a global workforce. Projects such as H3Africa1 and major biobank studies in several Asian countries2,3,4 have already provided crucial insights into genetic diversity and its links to human traits and diseases via genome-wide association studies (GWASs) carried out in cohorts more representative of global genetic diversity. However, much work remains to be done, as the Eurocentric bias in genetic research still prevails according to the GWAS Diversity Monitor: 95.2% European, 3.1% Asian, 0.2% African, 0.5% Afro-Caribbean/African American, 0.3% Hispanic/Latine, 0.7% other/mixed.5 Moreover, with research moving forward from GWASs to population functional genomic studies, representation bias remains a challenge. Addressing this issue is on the agenda of major research programs, funders, and many research groups.
While the lack of diversity in genetic and clinical studies has been widely discussed, there is an even more profound lack of genetic and environmental diversity in functional genomics datasets (Figure 1), which is the focus of this perspective and an issue only slowly starting to receive attention.6 Functional genomics data are used to understand the molecular processes that underlie human traits and bridge the gap between genotype and trait. Indeed, functional interpretation of GWASs has recently emerged as perhaps the biggest contemporary challenge in human genetics, as identifying molecular mediators of GWAS effects is necessary to translate GWAS discoveries into biological insights and medical benefits.
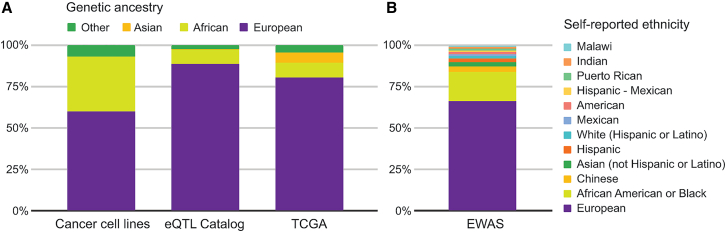
Figure 1.
Donor diversity in different functional genomics datasets and resources
(A and B) Genetic ancestry in a set of commonly used cancer cell lines,7 the eQTL Catalog,8 and TCGA9 are shown in (A), with (B) showing self-identified ethnicity across a variety of epigenome-wide association studies (EWASs).10 In addition to overrepresentation of samples of European ancestry and ethnicity, these data highlight the complexity of capturing the multiple dimensions of genetic ancestries, self-identities, and donor origins—information that is typically not collected and/or poorly reported in functional genomics data.
In contrast to well-established practices for metadata curation in GWAS repositories, functional genomic studies do not systematically identify ethnicity, ancestry, or environmental metadata of samples, hampering further analysis aiming to integrate this information. Despite efforts such as TOPMed and All of Us investing in more balanced multi-omic studies of minority populations in the United States as well as multiple national population biobanks, this alone will not fix gaps in global diversity in functional genomics research.11,12 While some of the underlying reasons are partially shared with those limiting the diversity of GWASs, such as the long legacy of research funding and capacity building in high-income countries (HICs),13 functional studies have additional unique challenges that we discuss in this perspective.
We argue for decisive efforts to address these bottlenecks to diversify functional genomics studies and realize the benefits of human genetics both for basic biological discovery and for human health worldwide. The imperative for remedying this bias is not simply moral. A focus on individuals leading predominantly sedentary lives in urbanized settings in HICs provides only a narrow representation of both extant human genetic diversity and the range of social, cultural, and physical environments occupied by humans. This significantly limits our collective ability to move beyond fundamental discovery to equitable clinical outcomes, as discussed further below.5,14
Importance of diversity in molecular studies
Two of the National Human Genome Research Institute’s ten “bold predictions” for 2030 for improving human health at the forefront of genomics are centered on diversity (genomic and workforce). The bold predictions also have a goal to go beyond historic social constructs such as race.15 However, lack of ancestral and environmental diversity in studies of molecular variation creates multiple roadblocks and challenges for GWAS discovery, interpretation, follow-up, and applications in precision medicine and predictive health (Figure 2). Firstly, most studies of molecular variation are performed in individuals living in urban or suburban environments that represent a narrow range of the environmental conditions in which humans live. Consequently, these studies may miss disease-relevant physiological variation and its molecular underpinnings, including host-pathogen interactions, gene-environment interactions in more extreme environments, societal stressors, and other factors.16,17,18,19 These molecular processes can include those also involved in disease processes, including chronic diseases that are not directly and immediately linked to environmental exposure, such as the role of infections in later autoimmune disease. With increasingly compelling evidence of the importance of such environmental exposures to human disease and their interactions with genetic risk,20,21,22 this represents a major gap in molecular studies and a missed opportunity in understanding diverse molecular triggers and risk factors of disease. This is not in disagreement with recent research on large cohorts in HICs suggesting that GWAS effect sizes are typically robust across ancestries,23 as environmental effects may be quite specific.
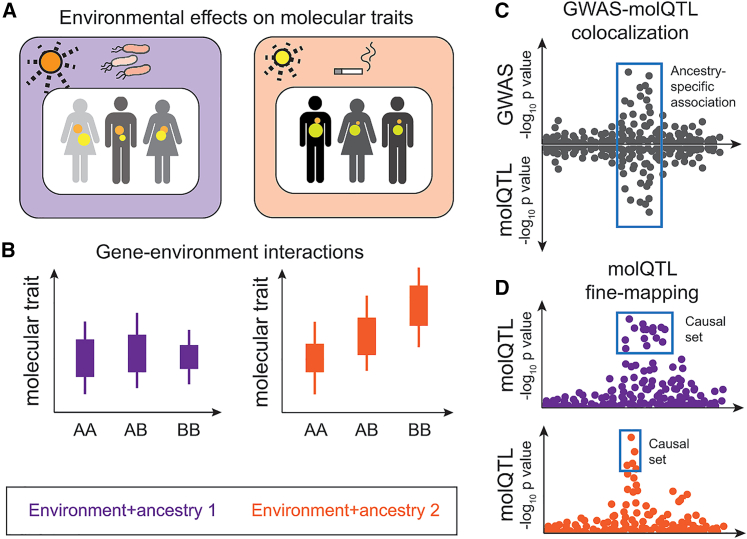
Figure 2.
Illustration of some of the advantages of diversity in functional genomics studies
(A) Capturing molecular traits and processes in tissues and cells (colored dots) that are associated to specific environmental exposures.
(B) Detecting gene-environment or gene-gene interactions.
(C) Enabling molecular QTL-GWAS colocalization including in loci that are present in only specific ancestries or environmental contexts.
(D) Improving fine-mapping by leveraging different linkage disequilibrium (LD) patterns in different ancestries, with the lower population having less LD in this locus and thus a smaller set of putative causal variants.
Secondly, one of the primary tools for characterizing potential molecular effects of noncoding GWAS loci is mapping their associations to molecular phenotypes (molecular quantitative trait locus [molQTL] mapping).24 While major molQTL resources have been assembled especially for expression and splicing QTLs (eQTLs and sQTLs, respectively), these are dominated by European ancestries and largely lack variants and haplotype patterns that are specific to other ancestries8 Thus, a simple in silico molQTL query is less fruitful for non-European GWAS loci, and the variant-to-function gap is thus wider for non-European GWAS loci due to the lack of data. This hampers our ability to use the growing GWAS datasets to learn about disease biology and to identify potential targets for drugs and other interventions. Furthermore, increased diversity of molQTL studies helps to break down linkage disequilibrium blocks, leading to better fine-mapping of likely causal regulatory variants.25,26 This is key to experimental follow-up, as discussed below, and may also help to increase trans-ancestry portability of polygenic risk scores.27
Studies of severe disease caused by rare variants—often with immediate clinical genomics applications—rely on large genetic databases of variant frequencies, and biases caused by lack of diversity are well recognized.28 Emerging multi-omic approaches for clinical genomics aim to use not only genomic data but also transcriptome, epigenome, and other data to support identification of disease-causing rare variants.29 These approaches are typically based on identifying population outliers in molecular profiles, such as situations where an individual has an aberrant splicing pattern in a gene where they carry a rare intronic variant. Lack of genetic variation data and molecular patterns that are an appropriate reference for individuals can give rise to false-positive diagnoses in individuals of non-European ancestries that may erroneously appear as being outside the general population spectrum. However, effects of population ancestry on molecular states and functions of cells should not be overstated: most of the biology at the molecular, cellular, tissue, and physiological levels is shared among all humans (and often also among many species). Furthermore, similarly to the partitioning of genetic variation,30,31 the majority of inter-individual variation in molecular traits is not explained by population or continental groupings.32 Thus, the lack of diversity and/or representativeness does not invalidate basic molecular biology discoveries, and the reasonable default assumption for most molecular processes is that they are broadly shared. Yet, in studies that aim to characterize basic molecular biology and typically analyze up to a handful of individuals as biological replicates, diverse sampling not only prevents biased assignments of what constitutes an “archetype” of humans but can also lead to more robust discoveries. When our attention is focused on variation between individuals, sample selection that includes genetic and environmental variation is key for the reasons described above, and the study of diverse populations can provide valuable insights into population-specific genetic risk factors. While the extent of variation in GWAS effect sizes due to the individual’s genetic and environmental background, and driven by epistatic and gene-environment interactions, is debated,23,33,34 it appears that effect-size heterogeneity is the exception rather than the rule. This offers promise for integration and portability of results across populations with careful statistical methods and empirical data from diverse populations, in a manner that does not necessarily require extremely large datasets from all populations, which would be a daunting challenge. Nevertheless, this recommendation should not be taken as justification to limit resources to increase sample sizes in studies focused on underrepresented populations, as samples from HICs do continue to be extensively analyzed.
Barriers and ways to fix them
Unlike DNA samples, which can be readily collected from blood or saliva and withstand substantial processing delays before nucleic acid isolation without impacting data quality, generation of high-quality functional genomics data is not just costly but also requires sample collection in a controlled environment, rapid initial processing, and special sample shipping conditions.35 This presents significant logistical challenges when working in low- and middle-income countries (LMICs), both in urban and non-urban environments, where facilities can be limited or under-resourced.36,37,38 Establishing local capacity for cost-effective functional genomic research and developing cohort studies and biobanks that actively engage local research communities, with the support of government, is critical in this regard.
Obtaining informed donor consent for biospecimen sampling without prior community engagement and buy-in is challenging and can be further complicated by a lack of trust in scientists. Varying cultural attitudes surrounding the handling of bodies after death, and toward invasive or postmortem biospecimen sampling introduce additional layers of complexity.39,40,41 It is important to have meaningful community involvement and engagement with ethical, legal, and social issues (ELSI) facilitators, which are fundamental to success in the long term (https://www.inegi.org.mx/programas/enpecyt/2017/). Ideally, this should be viewed not as an imposition, but as an opportunity for sustained inclusion and participation of community members at all stages of the project, including collaboration with local researchers and capacity building. Further opportunities for inclusion can be gained from innovation in less invasive biospecimen collection and study designs that accommodate different cultural contexts.42
Once consent has been secured and samples collected, the labile nature of RNA and other biomolecules of interest and their large dynamic range introduces additional difficulties. Small differences in sample processing across sites or handlers can introduce unwanted variability in datasets, making downstream integration and harmonization complex and time intensive. However, it is often impossible to process all samples in a central location. Beyond logistics, some countries have strict export bans on human biological samples because of past bio-colonialist, extractive research carried out by scientists based in HICs. Rather than a reason for exclusion of participants from LMICs, this should be seen as an opportunity to increase equity not only in sample collections but in the scientific teams generating them. In the short term, some consortia are making use of cross-site controls to aid in correcting batch effects, although even with these in place the analytical challenges are not to be underestimated. Complexity increases even more when the aim is to generate multiple modalities of molecular data, such as simultaneous assaying of gene expression and chromatin structure, or to integrate molecular data with other orthogonal types such as health records, which are not always interoperable across countries or health systems and, in any case, are much less likely to be digitized outside of HICs. Some groups have developed standards and then harmonized survey instruments across diverse LMICs and HICs to allow further metadata integration.43
Data generation and infrastructure building in LMICs is also hindered by higher costs for sequencing services and reagents, which due to lower demand are sometimes artificially inflated, a vicious cycle that hinders global scientific advancement.44,45 While international collaboration can be key for overcoming these challenges, these collaborations can be easily unbalanced in power. It is not uncommon for scientists in HICs to establish collaborations to access samples in LMICs without including as equal partners the scientists that collect the samples and that have been working with the local communities, excluding LMIC scientists from data analysis and discovery research stages. This is noticeable in the authorship positions of the articles resulting from these collaborations.46 Not including the perspective of LMIC scientists along the research process also hampers scientific development, as these perspectives could open new avenues for discovery. In this context, it is also important to highlight the tension between current discipline expectations for fully open and unrestricted data access and the growing recognition of Indigenous and national data sovereignty, e.g., as exemplified by the CARE principles.47,48 Here, the possibility of federating both data storage and analyses49 such that participants retain ongoing ownership of their data emerges as a solution worth exploring further. Additionally, peer review bias toward research developed in HICs50,51,52,53 slows down publication of work produced in LMICs and affects the availability of novel genomic data generated in these studies. Some journals, such as eLife and F1000, have proposed changes in the peer review process to increase accountability on the reviewers’ side to reduce unconscious bias and improve scientific input, but analyses remain to be done to probe if this strategy is yielding the desired results.
Governments and funders, too, have significant roles to play in bringing about change. Science spending in LMICs is generally low (https://www.oecd-ilibrary.org/science-and-technology/main-science-and-technology-indicators_2304277x), which hampers national capability to generate functional genomic data as grant budgets are limited. Fundamental science is not always a funding priority, especially considering more urgent medical needs.54,55,56 Few international funding agencies offer funding for research groups based in LMICs, especially for discovery-based research, and too often there is limited support for the preliminary outreach, community engagement, and logistic pilot studies that by necessity precede full-scale studies, because their small sample sizes are seen as uninformative. As exceptions to these trends, we highlight the Chan Zuckerberg Initiative’s Ancestry Networks for the Human Cell Atlas, which is supporting the generation of large single-cell-resolution datasets from countries across Africa, Asia, the Caribbean, and Latin America to incorporate into the Human Cell Atlas, as well as local outreach and community engagement, alongside the Wellcome Trust’s long-standing record of directly supporting fundamental research in LMICs. Nevertheless, these exceptional examples are not enough to shift the current tendency, and systematic funding opportunities in HIC funding agencies (private and governmental) should be made open to LMIC scientists. It is understandable that HIC funding agencies prioritize studies focused on their own populations; however, resource limitations in LMICs are, in many cases, a consequence of a colonial history. Making funding opportunities available could be a way of restorative justice to support research development.
We need funders in HICs to recognize the colonial debt with LMICs and allocate funds to support research development. Because this work can take many years to lead to results, it is also worth taking advantage of complementary approaches that can bear more immediate fruit. Amongst these we highlight the nascent potential of high-throughput multiplex assays of variant effect (MAVEs) or CRISPR-based perturbation screens to test the functional potential of tens of thousands of variants simultaneously.57,58 Adopting such approaches can aid in fine-mapping molQTLs or GWAS loci and in dissecting their molecular mechanisms regardless of the population where the variants and/or associations were originally detected. It is also possible to expose multiple cell lines to a defined set of external stimuli (e.g., pathogens, chemicals) and identify molQTLs associated with inter-individual response differences.59 However, except for lymphoblastoid cell lines from the 1000 Genomes Project,60 most public cell line collections are also highly biased toward European ancestries, perpetuating systematic exclusions of other populations.
Conclusions
Global challenges require global support. To increase our ability to improve and map genetic variation to health and disease, it is essential that we diversify across multiple variables, including genuine inclusion of LMICs. Beyond genomic ancestry, variables such as age, sex, environment, lifestyle, ethnicity, and country of origin, and the interactions between them, are important factors to incorporate into study designs to understand the influence of and interplay between sociology, geography, and biology. Inclusion of participants and scientists from diverse communities and the Global South will increase feasibility and improve studies targeting these populations to prevent helicopter science.61 In women, racial and ethnic minorities, and other underrepresented groups, we can disentangle and measure the influence of societal factors like social status (environment) on disease etiology and pathogenesis. The contribution of genomic ancestry is a modifier of the experiential, cultural factors that increase risk to disease and likely influence health. While there is much to be done to genuinely have an impact in diversifying functional genomics data generation, there is a major opportunity on multiple fronts to advance scientific discovery and global equity.
Acknowledgments
We would like to thank Charles Breeze for providing EWAS statistics of Figure 1B for re-plotting purposes. S.G. has grants from Chan Zuckerberg Initiative Ancestry Network (DAF2021-240624) and CDMRP Ovarian Cancer program (W81XWH-18-1-0072). Y.I. is supported by Chan Zuckerberg Initiative Ancestry Network grant DAF2022-239720. A.M.-R. is supported by Chan Zuckerberg Initiative Ancestry Network (2021-240438), CONACYT-FORDECYT-PRONACES grant numbers 11311 and 6390, and Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica–Universidad Nacional Autónoma de México (PAPIIT-UNAM) grant numbers IA203021 and IN218023. I.G.R. is supported by a Chan Zuckerberg Initiative Ancestry Network grant, Australian National Health and Medical Research Council Ideas grant 2020501, the European Union through Horizon 2020 Research and Innovation Program under grant number 810645, and the European Regional Development Fund project number MOBEC008.
Declaration of interests
S.G. has grants from Pfizer Inc. T.L. is a paid advisor or consultant to GSK, Pfizer, and Goldfinch Bio and has equity in Variant Bio.
Contributor Information
Sophia H.L. George, Email: sophia.george@med.miami.edu.
Tuuli Lappalainen, Email: tuuli.lappalainen@scilifelab.se.
References
- 1.Fatumo S., Inouye M. African genomes hold the key to accurate genetic risk prediction. Nat. Human Behav. 2023;7:295–296. doi: 10.1038/s41562-023-01549-1. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z., Chen J., Collins R., Guo Y., Peto R., Wu F., Li L., China Kadoorie Biobank CKB collaborative group China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int. J. Epidemiol. 2011;40:1652–1666. doi: 10.1093/ije/dyr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S., Jung P.E., Lee Y. Publicly-funded biobanks and networks in East Asia. SpringerPlus. 2016;5:1080. doi: 10.1186/s40064-016-2723-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagai A., Hirata M., Kamatani Y., Muto K., Matsuda K., Kiyohara Y., Ninomiya T., Tamakoshi A., Yamagata Z., Mushiroda T., et al. Overview of the BioBank Japan Project: Study design and profile. J. Epidemiol. 2017;27 doi: 10.1016/j.je.2016.12.005. S2–s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mills M.C., Rahal C. The GWAS Diversity Monitor tracks diversity by disease in real time. Nat. Genet. 2020;52:242–243. doi: 10.1038/s41588-020-0580-y. [DOI] [PubMed] [Google Scholar]
- 6.Breeze C.E., Beck S., Berndt S.I., Franceschini N. The missing diversity in human epigenomic studies. Nat. Genet. 2022;54:737–739. doi: 10.1038/s41588-022-01081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooker S.E., Jr., Woods-Burnham L., Bathina M., Lloyd S., Gorjala P., Mitra R., Nonn L., Kimbro K.S., Kittles R.A. Genetic Ancestry Analysis Reveals Misclassification of Commonly Used Cancer Cell Lines. Cancer Epidemiol. Biomarkers Prev. 2019;28:1003–1009. doi: 10.1158/1055-9965.EPI-18-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerimov N., Hayhurst J.D., Peikova K., Manning J.R., Walter P., Kolberg L., Samoviča M., Sakthivel M.P., Kuzmin I., Trevanion S.J., et al. A compendium of uniformly processed human gene expression and splicing quantitative trait loci. Nat. Genet. 2021;53:1290–1299. doi: 10.1038/s41588-021-00924-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan J., Hu Z., Mahal B.A., Zhao S.D., Kensler K.H., Pi J., Hu X., Zhang Y., Wang Y., Jiang J., et al. Integrated Analysis of Genetic Ancestry and Genomic Alterations across Cancers. Cancer Cell. 2018;34:549–560.e9. doi: 10.1016/j.ccell.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breeze C.E., Wong J.Y.Y., Beck S., Berndt S.I., Franceschini N. Diversity in EWAS: current state, challenges, and solutions. Genome Med. 2022;14:71. doi: 10.1186/s13073-022-01065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stark Z., Dolman L., Manolio T.A., Ozenberger B., Hill S.L., Caulfied M.J., Levy Y., Glazer D., Wilson J., Lawler M., et al. Integrating Genomics into Healthcare: A Global Responsibility. Am. J. Hum. Genet. 2019;104:13–20. doi: 10.1016/j.ajhg.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazareva T.E., Barbitoff Y.A., Changalidis A.I., Tkachenko A.A., Maksiutenko E.M., Nasykhova Y.A., Glotov A.S. Biobanking as a Tool for Genomic Research: From Allele Frequencies to Cross-Ancestry Association Studies. J. Personalized Med. 2022;12 doi: 10.3390/jpm12122040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirugo G., Williams S.M., Tishkoff S.A. The Missing Diversity in Human Genetic Studies. Cell. 2019;177:1080–1131. doi: 10.1016/j.cell.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 14.Ruan Y., Lin Y.F., Feng Y.C.A., Chen C.Y., Lam M., Guo Z., Stanley Global Asia Initiatives. He L., Sawa A., Martin A.R., et al. Improving polygenic prediction in ancestrally diverse populations. Nat. Genet. 2022;54:573–580. doi: 10.1038/s41588-022-01054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green E.D., Gunter C., Biesecker L.G., Di Francesco V., Easter C.L., Feingold E.A., Felsenfeld A.L., Kaufman D.J., Ostrander E.A., Pavan W.J., et al. Strategic vision for improving human health at The Forefront of Genomics. Nature. 2020;586:683–692. doi: 10.1038/s41586-020-2817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Idaghdour Y., Czika W., Shianna K.V., Lee S.H., Visscher P.M., Martin H.C., Miclaus K., Jadallah S.J., Goldstein D.B., Wolfinger R.D., Gibson G. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat. Genet. 2010;42:62–67. doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Favé M.J., Lamaze F.C., Soave D., Hodgkinson A., Gauvin H., Bruat V., Grenier J.-C., Gbeha E., Skead K., Smargiassi A., et al. Gene-by-environment interactions in urban populations modulate risk phenotypes. Nat. Commun. 2018;9:827. doi: 10.1038/s41467-018-03202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lea A., Subramaniam M., Ko A., Lehtimäki T., Raitoharju E., Kähönen M., Seppälä I., Mononen N., Raitakari O.T., Ala-Korpela M., et al. Genetic and environmental perturbations lead to regulatory decoherence. Elife. 2019;8 doi: 10.7554/eLife.40538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temba G.S., Kullaya V., Pecht T., Mmbaga B.T., Aschenbrenner A.C., Ulas T., Kibiki G., Lyamuya F., Boahen C.K., Kumar V., et al. Urban living in healthy Tanzanians is associated with an inflammatory status driven by dietary and metabolic changes. Nat. Immunol. 2021;22:287–300. doi: 10.1038/s41590-021-00867-8. [DOI] [PubMed] [Google Scholar]
- 20.Laville V., Majarian T., Sung Y.J., Schwander K., Feitosa M.F., Chasman D.I., Bentley A.R., Rotimi C.N., Cupples L.A., de Vries P.S., et al. Gene-lifestyle interactions in the genomics of human complex traits. Eur. J. Hum. Genet. 2022;30:730–739. doi: 10.1038/s41431-022-01045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virolainen S.J., VonHandorf A., Viel K.C.M.F., Weirauch M.T., Kottyan L.C. Gene–environment interactions and their impact on human health. Gene Immun. 2023;24:1–11. doi: 10.1038/s41435-022-00192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H., Eckhardt C.M., Baccarelli A.A. Molecular mechanisms of environmental exposures and human disease. Nat. Rev. Genet. 2023;24:332–344. doi: 10.1038/s41576-022-00569-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu S., Ferreira L.A.F., Shi S., Hellenthal G., Marchini J., Lawson D.J., Myers S.R. Leveraging fine-scale population structure reveals conservation in genetic effect sizes between human populations across a range of human phenotypes. bioRxiv. 2023 Preprint at. [Google Scholar]
- 24.Aguet F., Alasoo K., Li Y.I., Battle A., Im H.K., Montgomery S.B., Lappalainen T. Molecular quantitative trait loci. Nat. Rev. Methods Primers. 2023;3:4. [Google Scholar]
- 25.Vandiedonck C. Genetic association of molecular traits: A help to identify causative variants in complex diseases. Clin. Genet. 2018;93:520–532. doi: 10.1111/cge.13187. [DOI] [PubMed] [Google Scholar]
- 26.Uffelmann E., Huang Q.Q., Munung N.S., de Vries J., Okada Y., Martin A.R., Martin H.C., Lappalainen T., Posthuma D. Genome-wide association studies. Nat. Rev. Methods Primers. 2021;1:59. [Google Scholar]
- 27.Amariuta T., Ishigaki K., Sugishita H., Ohta T., Koido M., Dey K.K., Matsuda K., Murakami Y., Price A.L., Kawakami E., et al. Improving the trans-ancestry portability of polygenic risk scores by prioritizing variants in predicted cell-type-specific regulatory elements. Nat. Genet. 2020;52:1346–1354. doi: 10.1038/s41588-020-00740-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George S.H.L., Donenberg T., Alexis C., DeGennaro V., Jr., Dyer H., Yin S., Ali J., Butler R., Chin S.N., Curling D., et al. Gene Sequencing for Pathogenic Variants Among Adults With Breast and Ovarian Cancer in the Caribbean. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R.-S., Maron B.A., Loscalzo J. Multiomics Network Medicine Approaches to Precision Medicine and Therapeutics in Cardiovascular Diseases. Arterioscler. Thromb. Vasc. Biol. 2023;43:493–503. doi: 10.1161/ATVBAHA.122.318731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg N.A., Mahajan S., Ramachandran S., Zhao C., Pritchard J.K., Feldman M.W. Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg N.A. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann. Hum. Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- 32.Stranger B.E., Montgomery S.B., Dimas A.S., Parts L., Stegle O., Ingle C.E., Sekowska M., Smith G.D., Evans D., Gutierrez-Arcelus M., et al. Patterns of Cis Regulatory Variation in Diverse Human Populations. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdellaoui A., Dolan C.V., Verweij K.J.H., Nivard M.G. Gene–environment correlations across geographic regions affect genome-wide association studies. Nat. Genet. 2022;54:1345–1354. doi: 10.1038/s41588-022-01158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel R.A., Musharoff S.A., Spence J.P., Pimentel H., Tcheandjieu C., Mostafavi H., Sinnott-Armstrong N., Clarke S.L., Smith C.J., et al. VA Million Veteran Program Genetic interactions drive heterogeneity in causal variant effect sizes for gene expression and complex traits. Am. J. Hum. Genet. 2022;109:1286–1297. doi: 10.1016/j.ajhg.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallego Romero I., Pai A.A., Tung J., Gilad Y. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol. 2014;12:42. doi: 10.1186/1741-7007-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdrabou W., Dieng M.M., Diawara A., Sermé S.S., Almojil D., Sombié S., Henry N.B., Kargougou D., Manikandan V., Soulama I., Idaghdour Y. Metabolome modulation of the host adaptive immunity in human malaria. Nat. Metab. 2021;3:1001–1016. doi: 10.1038/s42255-021-00404-9. [DOI] [PubMed] [Google Scholar]
- 37.Suwalowska H. The invisible body work of 'last responders' - ethical and social issues faced by the pathologists in the Global South. Global Publ. Health. 2022;17:4183–4194. doi: 10.1080/17441692.2022.2076896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lea A.J., Garcia A., Arevalo J., Ayroles J.F., Buetow K., Cole S.W., Eid Rodriguez D., Gutierrez M., Highland H.M., Hooper P.L., et al. Natural selection of immune and metabolic genes associated with health in two lowland Bolivian populations. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2207544120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben Taher M., Pearson J., Cohen M., Offiah A.C. Acceptability of post-mortem imaging among Muslim and non-Muslim communities. Br. J. Radiol. 2018;91 doi: 10.1259/bjr.20180295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das M.K., Arora N.K., Debata P., Chellani H., Rasaily R., Gaikwad H., Meena K.R., Kaur G., Malik P., Joshi S., Kumari M. Why parents agree or disagree for minimally invasive tissue sampling (MITS) to identify causes of death in under-five children and stillbirth in North India: a qualitative study. BMC Pediatr. 2021;21:513. doi: 10.1186/s12887-021-02993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selket K., Glover M., Palmer S. Normalising post-mortems – whose cultural imperative? An indigenous view on New Zealand post-mortem policy. Kotuitui. 2015;10:1–9. [Google Scholar]
- 42.Martorella M., Kasela S., Garcia-Flores R., Gokden A., Castel S.E., Lappalainen T. Evaluation of noninvasive biospecimens for transcriptome studies. bioRxiv. 2023 doi: 10.1186/s12864-023-09875-4. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Odedina F.T., Ragin C., Martin D., Moser R., J.,O., McDonald A., Rice E.L., Nguyen J., Chinegwundoh F., Morisson Blidgen B., et al. Cancer Health Disparities; 2019. Standardized Global Behavioral and Epidemiological Measures for Prostate Cancer Studies in Black Men; pp. e1–e16. [Google Scholar]
- 44.Ciocca D.R., Delgado G. The reality of scientific research in Latin America; an insider's perspective. Cell Stress Chaperones. 2017;22:847–852. doi: 10.1007/s12192-017-0815-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olufadewa I.I., Adesina M.A., Ayorinde T. From Africa to the World: Reimagining Africa's research capacity and culture in the global knowledge economy. J. Glob. Health. 2020;10 doi: 10.7189/jogh.10.010321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Odeny B., Bosurgi R. Time to end parachute science. PLoS Med. 2022;19 doi: 10.1371/journal.pmed.1004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jennings L., Anderson T., Martinez A., Sterling R., Chavez D.D., Garba I., Hudson M., Garrison N.A., Carroll S.R. Applying the 'CARE Principles for Indigenous Data Governance' to ecology and biodiversity research. Nat. Ecol. Evol. 2023;7:1547–1551. doi: 10.1038/s41559-023-02161-2. [DOI] [PubMed] [Google Scholar]
- 48.Carroll S.R., Garba I., Figueroa-Rodríguez O.L., Holbrook J., Lovett R., Materechera S., Parsons M., Raseroka K., Rodriguez-Lonebear D., Rowe R., et al. The CARE Principles for Indigenous Data Governance. Data Sci. J. 2020;19:12. [Google Scholar]
- 49.Boscarino N., Cartwright R.A., Fox K., Tsosie K.S. Federated learning and Indigenous genomic data sovereignty. Nat. Mach. Intell. 2022;4:909–911. doi: 10.1038/s42256-022-00551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris M., Macinko J., Jimenez G., Mullachery P. Measuring the bias against low-income country research: an Implicit Association Test. Glob. Health. 2017;13:80. doi: 10.1186/s12992-017-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris M., Marti J., Watt H., Bhatti Y., Macinko J., Darzi A.W. Explicit Bias Toward High-Income-Country Research: A Randomized, Blinded, Crossover Experiment Of English Clinicians. Health Aff. 2017;36:1997–2004. doi: 10.1377/hlthaff.2017.0773. [DOI] [PubMed] [Google Scholar]
- 52.Fox C.W., Meyer J., Aimé E. Double-blind peer review affects reviewer ratings and editor decisions at an ecology journal. Functional Ecology. 2023;37:1144–1157. [Google Scholar]
- 53.Liu F., Rahwan T., AlShebli B. Non-White scientists appear on fewer editorial boards, spend more time under review, and receive fewer citations. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2215324120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pramesh C.S., Badwe R.A., Bhoo-Pathy N., Booth C.M., Chinnaswamy G., Dare A.J., de Andrade V.P., Hunter D.J., Gopal S., Gospodarowicz M., et al. Priorities for cancer research in low- and middle-income countries: a global perspective. Nat. Med. 2022;28:649–657. doi: 10.1038/s41591-022-01738-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pramesh C.S., Venkataramanan R., Suvarna V., Goel N.S., Lakshman S., Venkatesh V., Gupta V., Badwe R. Involvement of general public in biomedical research. Perspect. Clin. Res. 2016;7:152–155. doi: 10.4103/2229-3485.192029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charani E., Abimbola S., Pai M., Adeyi O., Mendelson M., Laxminarayan R., Rasheed M.A. Funders: The missing link in equitable global health research? PLOS Glob. Public Health. 2022;2 doi: 10.1371/journal.pgph.0000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Starita L.M., Ahituv N., Dunham M.J., Kitzman J.O., Roth F.P., Seelig G., Shendure J., Fowler D.M. Variant Interpretation: Functional Assays to the Rescue. Am. J. Hum. Genet. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morris J.A., Caragine C., Daniloski Z., Domingo J., Barry T., Lu L., Davis K., Ziosi M., Glinos D.A., Hao S., et al. Discovery of target genes and pathways at GWAS loci by pooled single-cell CRISPR screens. Science. 2023;380 doi: 10.1126/science.adh7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Findley A.S., Monziani A., Richards A.L., Rhodes K., Ward M.C., Kalita C.A., Alazizi A., Pazokitoroudi A., Sankararaman S., Wen X., et al. Functional dynamic genetic effects on gene regulation are specific to particular cell types and environmental conditions. Elife. 2021;10 doi: 10.7554/eLife.67077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Atutornu J., Milne R., Costa A., Patch C., Middleton A. Towards equitable and trustworthy genomics research. EBioMedicine. 2022;76 doi: 10.1016/j.ebiom.2022.103879. [DOI] [PMC free article] [PubMed] [Google Scholar]