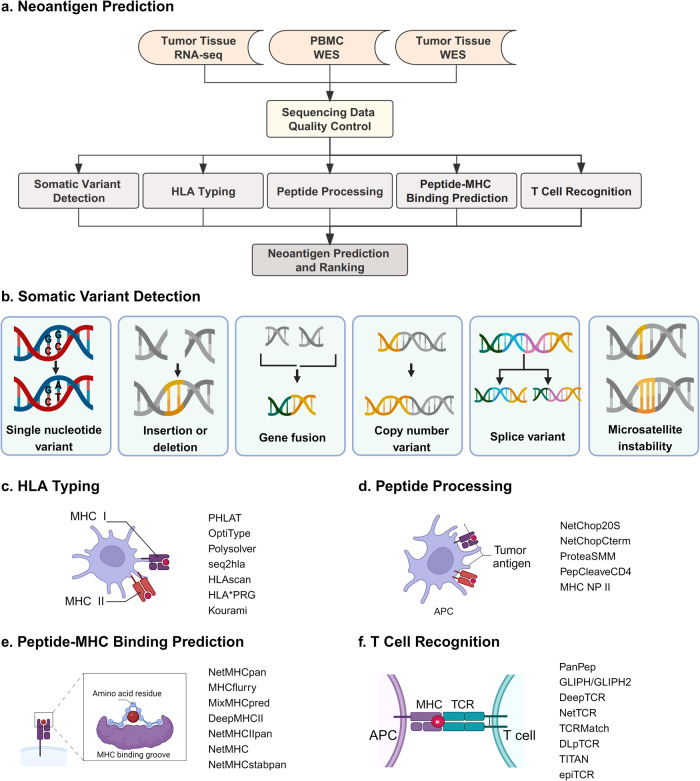
Fig. 3.
Prediction of neoantigen candidates. a An overview of the bioinformatic characterization of neoantigens. b The somatic mutants originate from multiple sources, including single nucleotide variants, insertions/deletions, gene fusion, copy number variant, splice variant and microsatellite instability. c HLA typing from WES and RNA-seq could be found by the in-silico tools. d Peptide processing prediction with several algorithms. e Available bioinformatic pipelines for peptide-MHC binding prediction. f Available bioinformatic pipelines for T cell recognition. RNA-seq, RNA-sequencing; WES Whole exome sequencing, PBMC Peripheral blood mononuclear cell, HLA Human leukocyte antigen, MHC Major histocompatibility complex, APC Antigen presentation cell, TCR T cell receptor