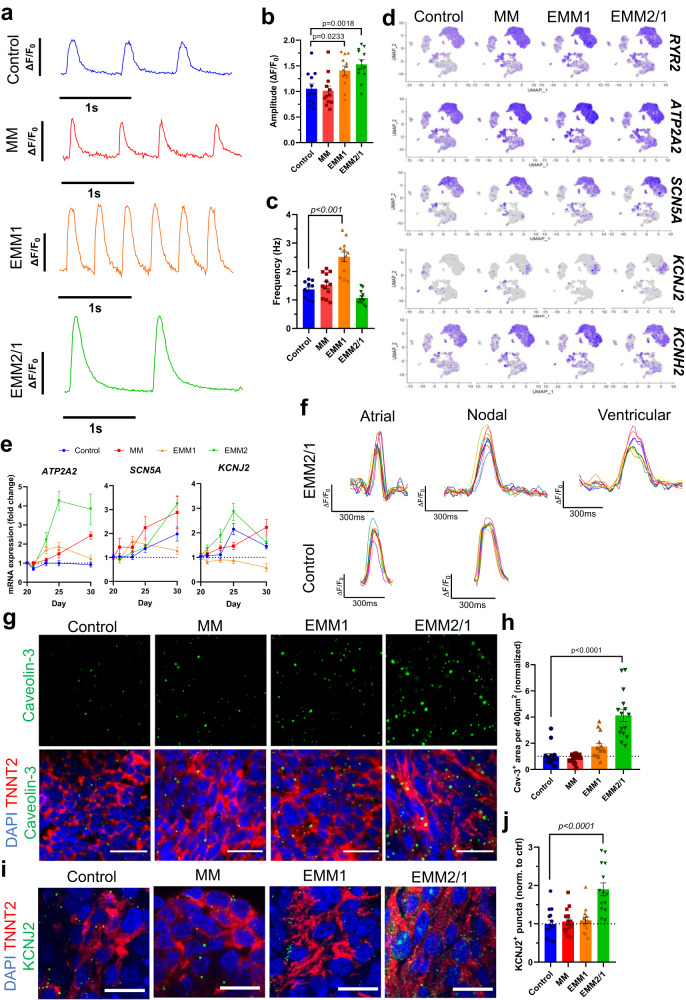
Fig. 5. Developmental induction conditions promote progressive electrophysiological maturation in human heart organoids.
a Representative calcium transient traces within d30 human heart organoids from each condition (n = 12 independent organoids per condition across three independent experiments). Traces represent data from an individual cardiomyocyte within human heart organoids. b Quantification of peak amplitude of calcium transient traces from each condition (n = 12 across three independent experiments). Values = mean ± s.e.m. c Quantification of calcium transient peak frequency for each condition (n = 12 independent organoids per condition across three independent experiments). Values = mean ± s.e.m. d Feature plots displaying key electrophysiological genes differentially expressed in the VCM and ACM clusters in each condition. e mRNA expression of key electrophysiological genes between d20 and d30 for each condition (n = 8 independent organoids per day per condition per gene across three independent experiments). Data presented as log2 fold change normalized to d20. Values = mean ± s.e.m. f Representative voltage tracings of organoids in the EMM2/1 and control conditions depicting atrial-, nodal-, and ventricular-like action potentials (n = 8 (EMM2/1, Ventricular), 9 (EMM2/1, Atrial and Nodal; Control, Atrial), and 10 (Control, Nodal) individual cells from three independent organoids across three independent experiments). Representative immunofluorescence images (g) and quantification (h) of caveolin-3 puncta for each condition (n = 15 independent organoids per condition across three independent experiments). Green = caveolin-3, red = TNNT2, blue = DAPI. Scale bar = 20 μm. Data presented as fold change normalized to control. Values = mean ± s.e.m., one-way ANOVA with Dunnett’s multiple comparisons tests. Representative immunofluorescence images (i) and quantification (j) of KCNJ2+ puncta for each condition (n = 14 independent organoids per condition across three independent experiments). KCNJ2 = green, TNNT2 = red, DAPI = blue. Scale bar = 20 μm. Data presented as fold change normalized to control. Values = mean ± s.e.m., one-way ANOVA with Dunnett’s multiple comparisons tests. Source data are provided as a Source Data file.