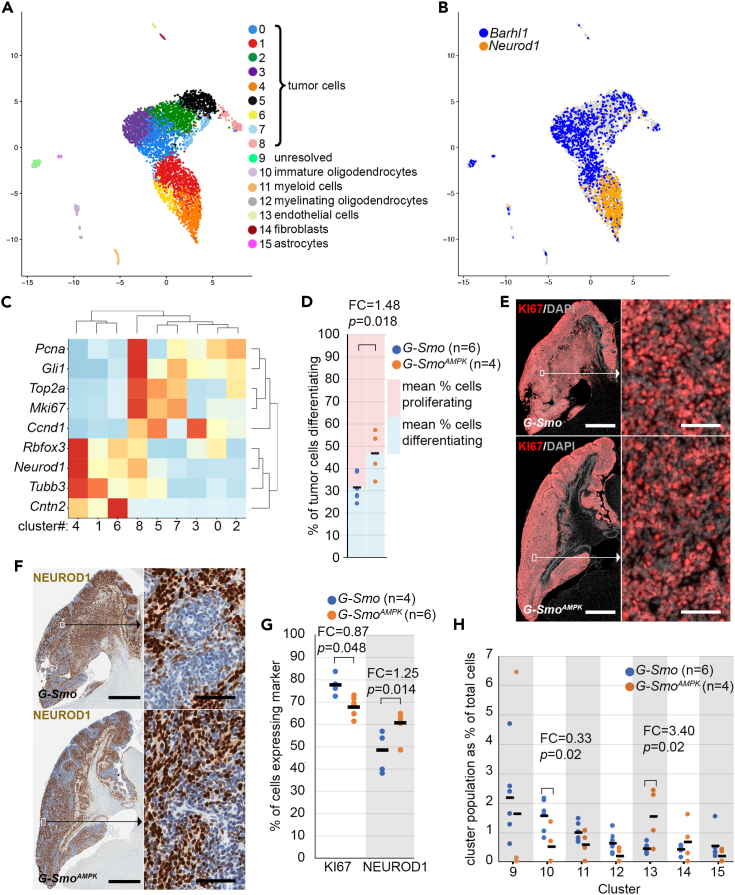
Figure 2.
AMPK inactivation alters cellular heterogeneity in SHH driven medulloblastomas
(A) UMAP plot of all cells from G-Smo and G-SmoAMPK tumors, color-coded by cluster. Cells are localized according to their proximity in PCA space.
(B) Feature plot showing the expression of Barhl1 and Neurod1, color coded over the UMAP shown in (A).
(C) Heatmap showing the scaled expression of indicated genes in each tumor cell cluster. Hierarchical clustering of genes appropriately grouped the proliferation markers and differentiation markers into two discrete sets. Hierarchical clustering of clusters 0–8 grouped the clusters into two discrete sets that corresponded with the expression of differentiation and proliferation markers.
(D) Comparison between genotypes of the fractions of differentiation tumor cells (Clusters 1, 4 and 6) from each replicate. The blue and red regions show the mean % differentiating and reciprocal % proliferating, indicating the balance between proliferation and differentiation within each genotype.
(E and F) Representative (E) KI67 IF and (F) NEUROD1 IHC, on sagittal sections of G-Smo and G-SmoAMPK medulloblastomas.
(G) Comparison of the fractions of tumor cells from each replicate expressing KI67 and NEUROD1.
(H) Comparison of populations of each stromal cluster in G-Smo and G-SmoAMPK tumors. In panels (D), (G) and (H), horizontal bars indicate the mean values for the conditions, dots represent values for individual replicates, and the number of visible dots may be smaller than the sample size (n) due to multiple replicates showing similar values. In (D) and (G), Student’s t test was used to make pairwise comparisons. In (H), Dirichlet regression was used to make statistical comparisons. In panels (E) and (F), bars = 1 mm in the left panel and 25 μm in the right panel.