Figure 2.
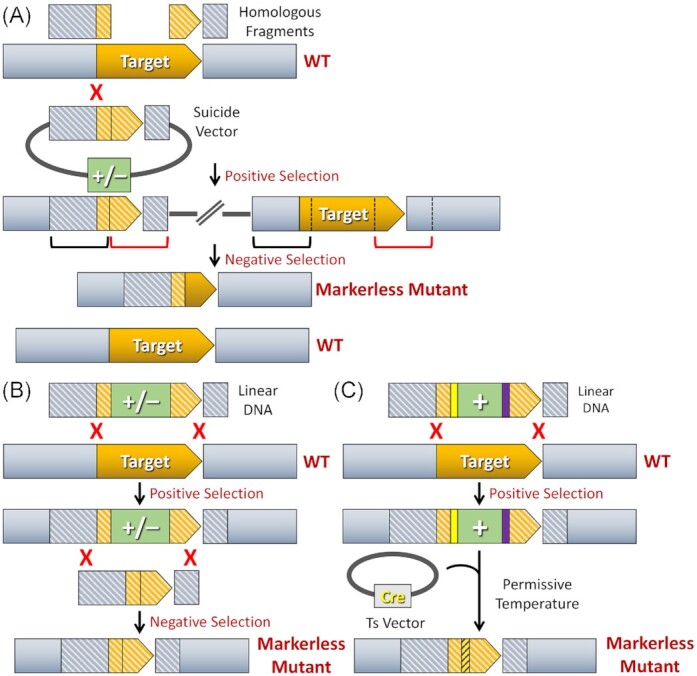
Markerless mutagenesis strategies. (A) Counterselection with insertion duplication mutagenesis. Two equally sized homologous fragments (illustrated in grey and orange stripes) flanking the intended mutation site are cloned adjacent to each other on a suicide vector containing both positive and negative selection markers (‘+/–’; illustrated in green). Following transformation of the construct, one of the two fragments will randomly insert to the chromosome via single crossover homologous recombination. The same final outcome is achieved irrespective of which of the two homologous fragments recombines. Therefore, only one option is illustrated. Since the suicide vector contains two homologous fragments, both of these segments will be duplicated on the chromosome after the plasmid has inserted (as indicated by the red and black brackets). Negative selection is used to isolate clones in which these homologous segments have randomly recombined to excise the inserted vector from the chromosome. In this example, a markerless mutant will be created following a recombination event between the homologous segments marked by red brackets. However, if recombination occurs between the homologous segments marked by black brackets, a wild-type genotype will result. Consequently, counterselection with insertion duplication mutagenesis yields a mixed population of clones consisting of 50% mutant and 50% wild-type genotypes. (B) Counterselection with allelic replacement mutagenesis. Two homologous fragments flanking the intended mutation site of the target gene are ligated to the corresponding 5′ and 3′ ends of a counterselection cassette (‘+/–’; illustrated in green). Following transformation of this construct, both homologous fragments recombine with the chromosome, which deletes all of the intervening chromosomal DNA between the homologous segments and replaces it with the counterselection cassette. The resulting strain is then transformed with an unmarked mutagenesis construct and subjected to negative selection to isolate the double crossover recombinants that have deleted the counterselection cassette. All of the resulting transformants should contain the desired markerless mutant genotype. (C) Recombinase-mediated markerless mutagenesis. A typical double crossover allelic replacement construct is assembled using a positive selection cassette (‘+’; illustrated in green) flanked by two Cre recombinase-dependent loxP sites (illustrated in yellow and purple). The allelic replacement mutant is next transformed with a temperature sensitive plasmid encoding the cre gene. Growth at the permissive temperature supports plasmid replication and ectopic production of the Cre recombinase. After a predetermined number of generations, the cells are shifted to the non-permissive temperature to trigger loss of the temperature sensitive cre expression plasmid. Plasmid-free clones are finally screened to identify those that have undergone Cre-mediated excision of the antibiotic cassette. Strains exhibiting the markerless mutant genotype will also retain a hybrid loxP site (illustrated in yellow and purple stripes) created via Cre-mediated recombination between the two original loxP sites.
