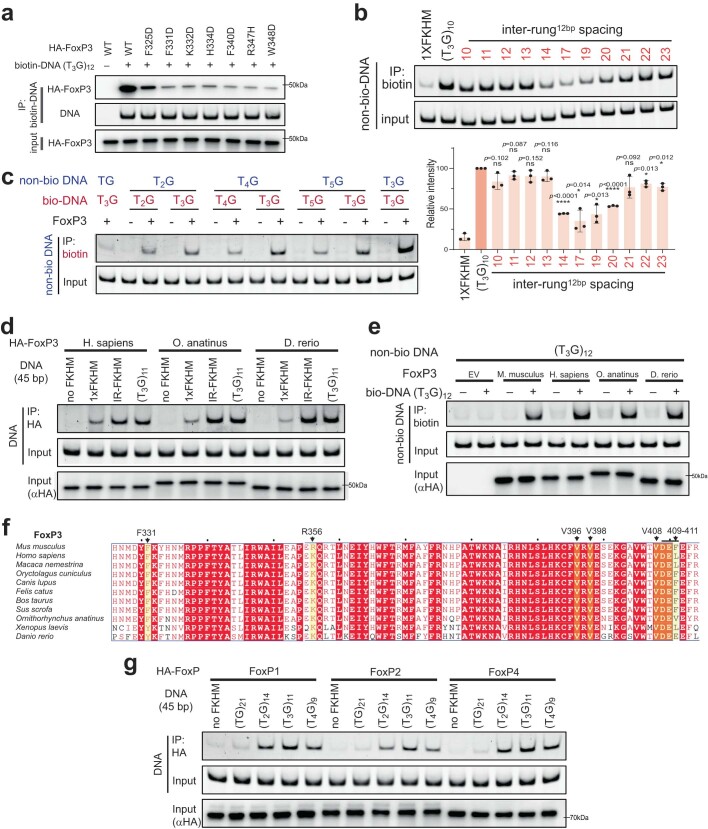
Extended Data Fig. 6. Multimerization on TnG repeats is conserved in FoxP3 orthologs and paralogs.
a. TnG repeats DNA-binding activity of FoxP3 with mutations in RBR. All seven RBR mutations previously shown to disrupt the head-to-head dimerization22 disrupted (T3G)12 binding. b. Effect of inter-rung12bp spacing variations on FoxP3-mediated DNA bridging. Non-biotinylated DNAs in Fig. 4d were mixed with biotinylated (T3G)10 and FoxP3∆N (0.2 μM) prior to streptavidin pull-down and gel analysis. Relative level of non-biotinylated DNA co-purified with biotinylated DNA was quantitated from three independent pull-downs. Two-tailed paired t-tests, in comparison to (T3G)10. p < 0.001 for ***, p < 0.05 for * and p > 0.05 for ns. c. DNA-bridging activity of FoxP3 with different combinations of TnG repeats. Biotinylated-DNA (red) and non-biotinylated DNA (blue) were mixed at 1:1 ratio and were incubated with FoxP3∆N (0.2 μM) prior to streptavidin pull-down and gel analysis of non-biotinylated DNA. T2G, T4G and T5G repeats bridged better with T3G repeats than with themselves. d. DNA-binding activity of FoxP3 orthologs with indicated DNA. Experiments were performed as in Fig. 5b. e. DNA-bridging activity of FoxP3 orthologs. Experiments were performed as in Fig. 5c. f. Sequence alignment of FoxP3 orthologs from different species, showing conservation of the key interface residues (yellow highlight, arrows on top with the residue identities in M. musculus FoxP3). g. DNA-binding activity of FoxP3 paralogs with TnG repeats (n = 1-4). Experiments were performed as in Fig. 5b.