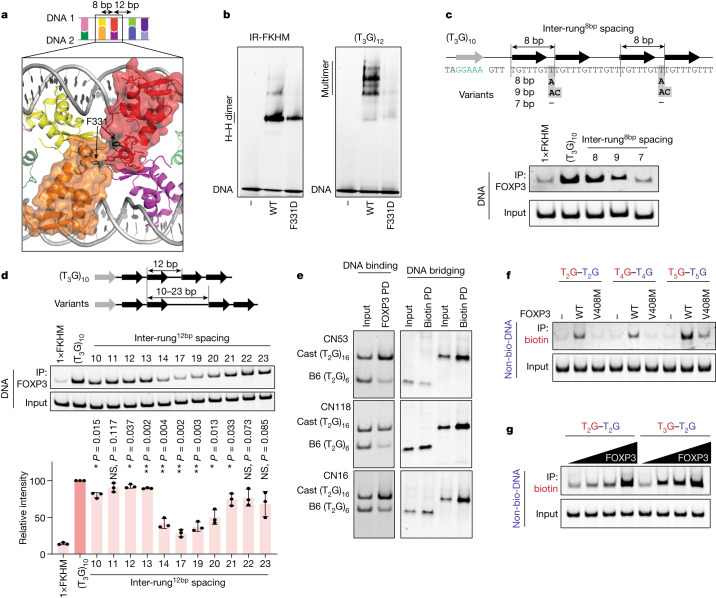
Fig. 4. Architectural flexibility of the ladder-like assembly broadens the sequence specificity of FOXP3.
a, Structure highlighting the inter-rung8bp interactions between the orange and red subunits (in surface representation). The interactions are primarily between RBRs, where Phe331 resides. b, The effect of the inter-rung8bp mutation (F331D) on the FOXP3–DNA interaction was analysed using FOXP3(∆N) pull-down. c, The effect of inter-rung8bp spacings on FOXP3 binding was determined using FOXP3(∆N) pull-down. Both inter-rung8bp gap nucleotides were changed from T in (T3G)10 to A (8 bp spacing), to AC (9 bp spacing) or to no nucleotide (7 bp spacing). The black arrows indicate FOXP3 footprints. The grey arrow and green-coloured nucleotides indicate the NFAT-binding site. NFAT interacts with FOXP3 and helps in fixing the FOXP3–DNA register, which was necessary to examine the effect of DNA sequence variations at or between the FOXP3 footprints. d, The effect of inter-rung12bp spacings on FOXP3 binding was analysed using FOXP3(∆N) pull-down. The inter-rung12bp spacing was changed from 12 bp in (T3G)10 to 10–23 bp (the sequences are provided in Supplementary Table 2b). The average recovery rate of DNA from three independent pull-downs was plotted. Statistical analysis was performed using two-tailed paired t-tests in comparison to (T3G)10; *P < 0.05; NS, P > 0.05. e, Comparison of Cast and B6 sequences in FOXP3 binding (left) and DNA bridging (right). Three pairs of sequences at the loci CN53, CN118 and CN16 with Cast bias in the CNR-seq analysis were compared (Supplementary Table 2a) using FOXP3(∆N) pull-down. f, DNA bridging between (T2G)14 and (T2G)14, between (T4G)9 and (T4G)9, and between (T5G)7 and (T5G)7 in the presence of WT FOXP3 or the IPEX mutant V408M. Biotinylated and non-biotinylated DNA are coloured red and blue, respectively. g, DNA bridging between (T2G)14 and (T2G)14, and between (T2G)14 and (T3G)11 by FOXP3 (0–0.4 μM).