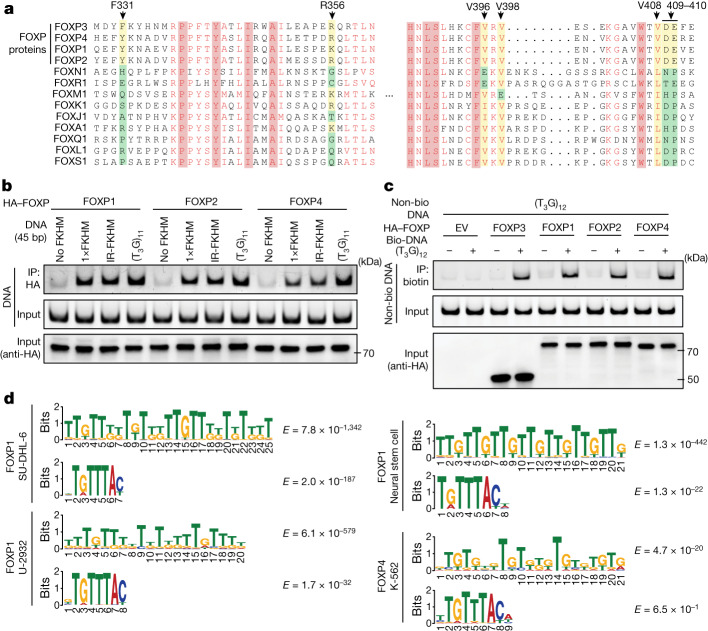
Fig. 5. TnG microsatellite recognition is conserved among FOXP3 orthologues and paralogues.
a, Sequence alignment of forkhead TFs. Residues equivalent to the key interface residues in mouse FOXP3 (arrow on top) were highlighted in yellow (when similar to the mouse residues) or green (when dissimilar). b, The DNA-binding activity of FOXP1, FOXP2 and FOXP4. HA-tagged FOXP1, FOXP2 and FOXP4 were transiently expressed in HEK293T cells and purified by anti-HA immunoprecipitation. Equivalent amounts of the indicated DNAs (all 45 bp) were added to FOXP1/2/4-bound beads and further purified before analysis using gel analysis (SybrGold). c, The DNA-bridging activity of FOXP1, FOXP2 and FOXP4. Experiments were performed as described in Fig. 3c using HEK293T lysate expressing HA-tagged FOXP TFs. d, De novo motif analysis of FOXP1 and FOXP4 ChIP–seq peaks from a published database38,39. The comprehensive list and their references are provided in Supplementary Table 1c.