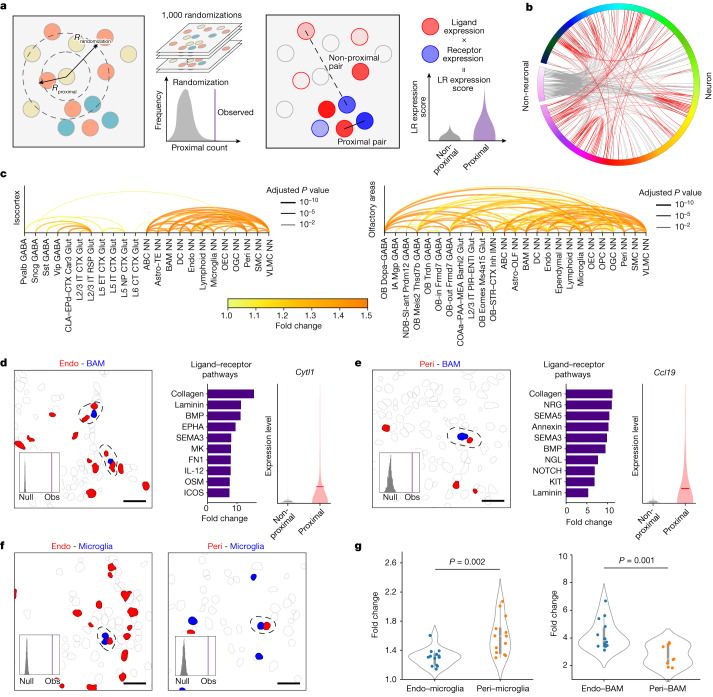
Fig. 6. Cell–cell interactions and communications.
a, Schematics of cell–cell interaction analysis (left) and ligand–receptor (LR) analysis (right). Rproximal denotes the proximity distance threshold; two cells are considered in contact or proximity if the distance between their centroid positions is within this distance threshold. Rrandomization denotes the randomization radius; we shifted the spatial location of each cell to a random position within R from its original location togenerate the null distribution. b, Cell–cell interactions across the whole brain. Each line corresponds to a predicted interacting cell-type pair. The grey lines indicate interactions between non-neuronal cells and neurons or among non-neuronal cells; the red lines indicate neuron–neuron interactions. c, Cell–cell interactions in two brain regions. Each line corresponds to an interacting cell-type pair, with the colour indicating fold change in proximity frequency compared with random chance and thickness indicating P values corrected by the Benjamini–Hochberg procedure. CLA, claustrum; CT, cortical-thalamic; EPd, endopiriform nucleus, dorsal part; ET, extratelencephalic; IT, intratelencephalic; L2/3, layer 2/3; NN, non-neuronal; NP, near-projecting. d, Interactions between endothelial cells and BAMs. Example image of cells, with cells of the indicated cell types shown in red and blue and all other cells shown in grey (left). Proximal cell pairs are circled by a dashed line. Observed counts (Obs) of the proximal cell pairs and the null distributions (null) from randomization control are shown in the inset. Top 10 ligand–receptor pathways upregulated in proximal cell pairs as compared to non-proximal cell pairs (middle). When multiple ligand–receptor pairs in a pathway are upregulated, the plotted fold-change value represents that of the pair with the highest upregulation fold change. Expression distributions of the indicated gene in endothelial cells proximal (red) or non-proximal (grey) to BAMs (right). Scale bar, 30 μm. Horizontal lines in the violin plots indicate median. e, Same as d, but for interactions between pericytes and BAMs. f, Interactions between endothelial cells and microglia (left) and between pericytes and microglia (right). g, Fold changes of observed proximal cell-pair number relative to the null-distribution mean across different brain regions. Each data point represents a brain region where significant interactions were observed (P values were calculated by two-sided Welch’s t-test; the centre points indicate the median, the boxes denote the interquartile range and the whiskers indicate 1.5 times the interquartile range). Comparison between endothelial–microglia and pericyte–microglia interactions (left), and comparison between endothelial–BAM and pericyte–BAM interactions (right).