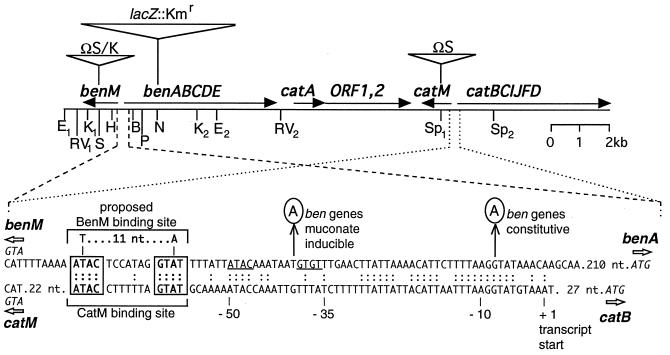
FIG. 2.
Restriction map of the chromosomal ben-cat region. The locations of genes, and their transcriptional directions, are shown relative to some of the known restriction endonuclease sites: EcoRI (E), EcoRV (RV), KpnI (K), SalI (S), HindIII (H), BsaAI (B), PvuII (P), NsiI (N), and SphI (Sp). Triangles mark the insertion sites of ΩS (29), ΩK (6), and the lacZ::Kmr cassette (18) of strains listed in Table 1. Below the map, the DNA sequence of the benM-benA intergenic region was aligned with the corresponding catM-catB intergenic region, with identical nucleotides indicated (:). The known CatM binding site (30) was used to predict a possible BenM binding site that has the consensus sequence (T-11 nt-A) and dyad symmetry (ATAC, GTAT) involved in binding LysR-type activators. An adjacent region with similar sequences is underlined. Circled nucleotides indicate mutations identified in strains ACN147 and ACN149 (G→A) and in strain ACN146 (T→A).