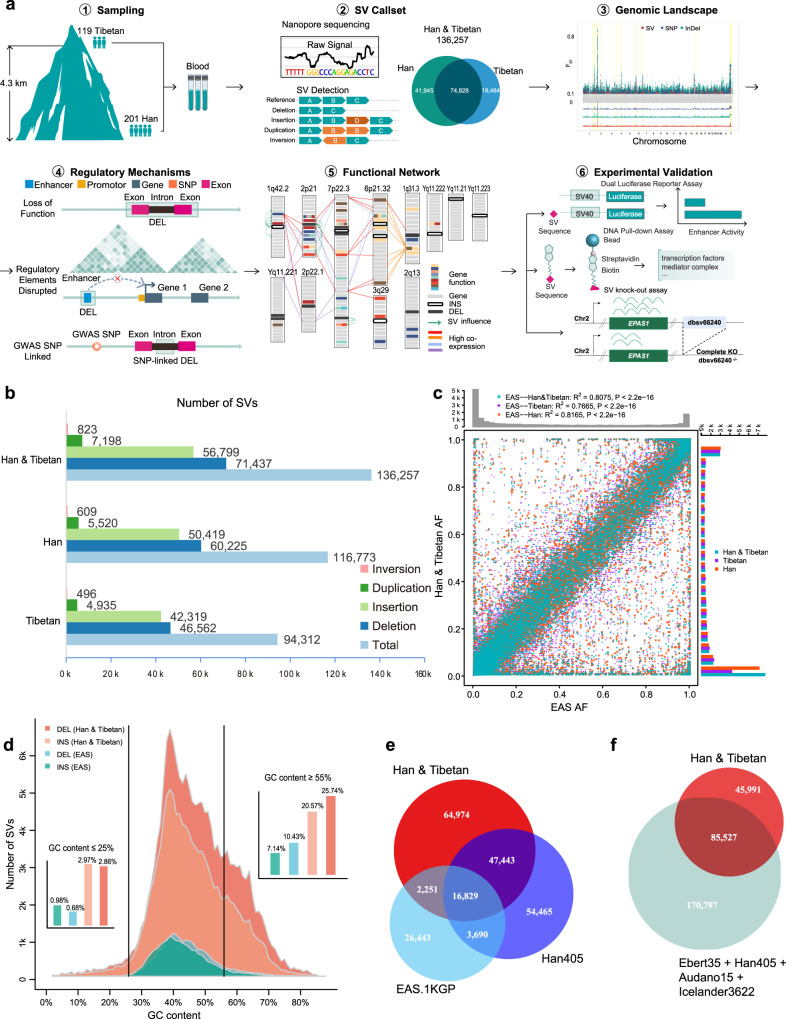
Fig. 1. SV discovery in 119 Tibetan and 201 Han samples.
a Summary of the experimental pipeline. Overall, 320 Han and Tibetan samples were collected and sequenced using the ONT platform, resulting in 136,257 SVs. Candidate SVs for high-altitude adaptation, their functional regulatory mechanisms based on their connections with exons, enhancers and TAD boundaries, and candidate genes for high-altitude adaptation were explored. Cis- and trans-regulatory circuitry rewiring was validated using three biological assays. b The numbers of 4 types of SVs in Han, Tibetan, and all samples. The majority are deletions and insertions. c Allele frequency consistency between the SVs in the Han and Tibetan cohort and the 1KGP EAS cohort. The high consistency between them indicates the high quality of our SV call set. d Distribution of the GC content in insertions and deletions in the Han and Tibetan cohort and the EAS cohort, indicating that the ONT platform performs well even in genomic regions with a biased GC content. e Overlap between the SVs of our cohort and EAS in the 1KGP and Han405 cohorts, showing a 43% increase with 64,974 novel SVs. f Overlap between the SVs of our cohort and other public long-read-sequencing-based SV call sets, consisting of Ebert35, Han405, Audano15 and Icelander3622, showing 45,991 novel SVs.