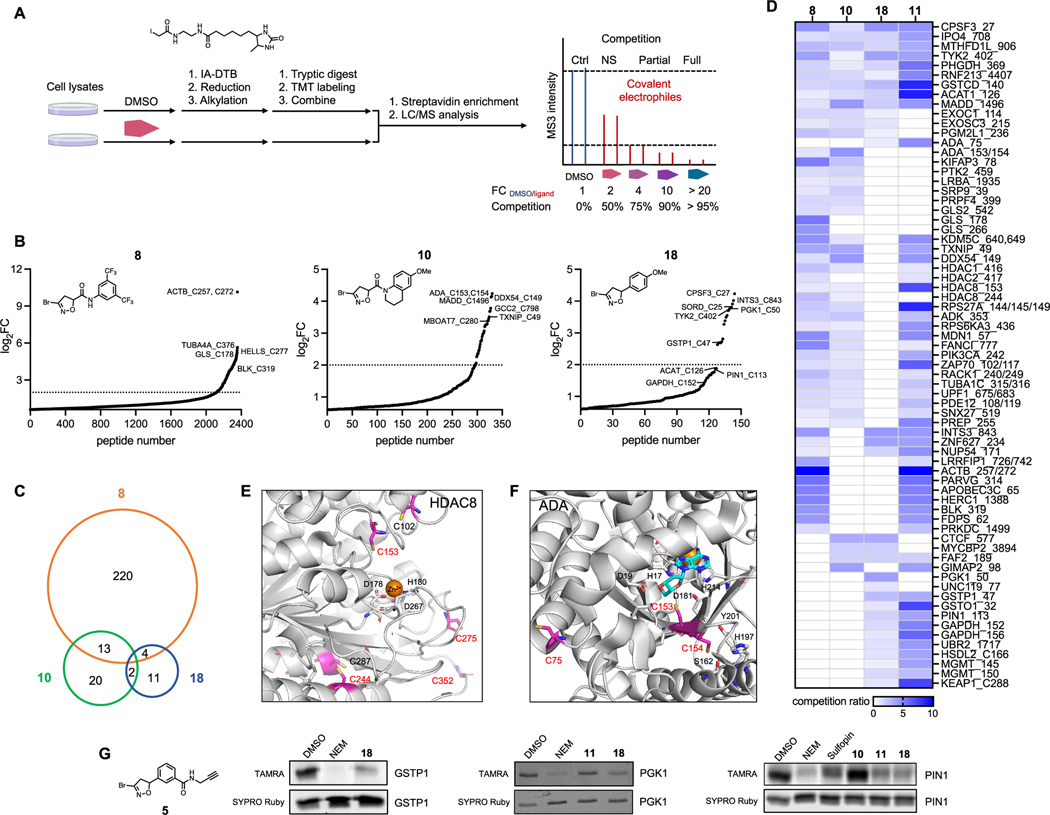
Figure 4.
Competitive MS-based reactivity profiling defines cysteines liganded by BDHI analogues. (A) Workflow for competitive measurement of cysteine ligandability with fragment electrophiles using IA−DTB probe. Steps include the treatment of cell lysate with compounds (500 μM, 4 h), treatment with IA−DTB probe (500 μM, 2 h), trypsin digestion, TMT labeling, combination, avidin enrichment for biotinylated cysteine-containing peptides, and analysis of competition ratio. (B) Structures and competition ratios (log2 FC values) from Jurkat cell proteome treated with BDHI 8, 10, and 18. (C) Venn diagram representation of the number of cysteine sites significantly liganded (log2 FC > 2) by BDHIs. Results were obtained by comparing the site overlap at a given competition ratio threshold for each ligand. (D) Heatmap of competition ratios for representative cysteines and fragments. (E) Crystal structure of HDAC8 (PDB: 2V5W). The zinc binding domain is shown along with reactive cysteines (depicted in magenta). (F) Crystal structure of substrate-bound, human ADA (PDB: 3IAR) with possible cysteine sites for covalent modification. Adenosine is depicted in cyan. (G) Competition binding assay between BDHI 18 and BDHI-alkyne 5. Recombinantly expressed and purified PGK1, GSTP1, and PIN1 were treated with each compound (500 μM, 4 h), labeled with BDHI 5 (500 μM, 2 h), and conjugated with TAMRA-N3 for in-gel fluorescence scanning.