Abstract
Background
Edible plants can exert anti-inflammatory activities in humans, being potentially useful in the treatment of inflammatory diseases. Plant-derived microRNAs have emerged as cross-kingdom gene expression regulators and could act as bioactive molecules involved in the beneficial effects of some edible plants. We investigated the role of edible plant-derived microRNAs in the modulation of pro-inflammatory human genes.
Methods
MicroRNAs from plant-derived foods were identified by next-generation sequencing. MicroRNAs with inflammatory putative targets were selected, after performing in silico analyses. The expression of candidate plant-derived miRNAs was analyzed by qPCR in edible plant-derived foods and their effects were evaluated in THP-1 monocytes differentiated to macrophages. The bioavailability of candidate plant miRNAs in humans was evaluated in feces and serum samples by qPCR.
Results
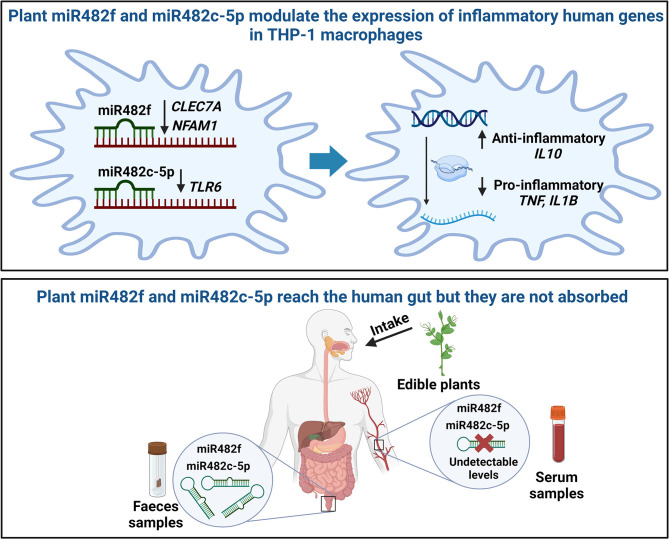
miR482f and miR482c-5p are present in several edible plant-derived foods, such as fruits, vegetables, and cooked legumes and cereals, and fats and oils. Transfections with miR482f and miR482c-5p mimics decreased the gene expression of CLEC7A and NFAM1, and TRL6, respectively, in human THP-1 monocytes differentiated to macrophages, which had an impact on gene expression profile of inflammatory biomarkers. Both microRNAs (miR482f and miR482c-5p) resisted degradation during digestion and were detected in human feces, although not in serum.
Conclusion
Our findings suggest that miR482f and miR482c-5p can promote an anti-inflammatory gene expression profile in human macrophages in vitro and their bioavailability in humans can be achieved through diet, but eventually restricted at the gut level.
Keywords: CLEC7A, NFAM1, TLR6, microRNA, cross-kingdom regulation, diet, inflammation, xenomiRs
Graphical Abstract
1 Introduction
Chronic inflammation is associated with the development of several comorbidities, such as cardiovascular diseases, cancer, autoimmune disorders, metabolic syndrome, type 2 diabetes, and non-alcoholic fatty liver disease, which altogether are the leading causes of mortality worldwide (1–4). Among the factors that could trigger long-term inflammation are obesity, chronic infections, gut microbiota dysbiosis, social, lifestyle, and environmental factors (1). The mechanisms underlying inflammation are complex and comprise several signaling pathways, including the nuclear factor kappa-B (NF-κB), mitogen-activated protein kinase (MAPK), and Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathways (5). Factors that could regulate these inflammatory pathways include Toll-like receptors (TLRs), microbial products, and pro-inflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β) (5).
Plant-based diets have been associated with the improvement of inflammation. Reported evidence has shown that consumption of nuts, fruits, vegetables, grains, and olive oil promotes an anti-inflammatory profile (6–11). However, the role of plant-based diets in inflammation in humans is currently under investigation; in particular, the identification of all bioactive molecules and their specific mechanisms of action. Among the plant bioactive compounds with anti-inflammatory activities are the phytochemicals, vitamins, minerals, and oil compounds, such as phenolics and triterpenoids found in fruits and vegetables; lectins and peptides in legumes; fiber, polyphenols, phytosterols, monounsaturated fatty acids (MUFAs), polyunsaturated fatty acids (PUFAs), vitamin E, selenium, and copper in nuts and peanuts; and polyphenols and fiber in cereals (6, 12–14). Notably, microRNAs (miRNAs) have recently emerged as new key bioactive molecules in plants (15, 16).
miRNAs are non-coding RNA molecules of approximately 22 nucleotides that exert post-transcriptional gene expression regulation by interacting with messenger RNAs (mRNAs) (17). Plant-derived miRNAs are crucial components of the biology of plants, playing a part in a wide range of functions, such as stress responses, development, and metabolism (18). Increasing evidence shows that edible plant-derived miRNAs could also modulate mammalian gene expression, influencing their physiology, for which they have become cross-kingdom gene expression regulators, and they stand for as therapeutic tools to treat human diseases (15, 16, 19, 20). Plant-derived miRNAs could modulate host metabolism, cell proliferation, apoptosis, and viral infections by interacting with diverse cell types, such as adipocytes, hepatocytes, enterocytes, and tumoral cells (21–27). However, there are controversial findings about the bioavailability of edible plant-derived miRNAs in the host (28). While some authors have found plant-derived miRNAs in animal tissues and blood samples, others have reported undetectable levels of these molecules in the host (29, 30). Further investigation is mandatory in order to clarify whether miRNAs could be relevant bioactive components of edible plants in modulating host physiology.
The general aim of the present study was to identify miRNAs in edible plant-based foods that could regulate the expression of human genes associated with inflammation. For this purpose, we selected several food types, including fruits, vegetables, legumes, cereals, and fats and oils, that have been previously associated with beneficial anti-inflammatory effects (6–11). The first objective consisted of identifying miRNAs in edible plant-based products that could display anti-inflammatory effects through modulation of human genes, by performing next-generation sequencing (NGS) and bioinformatic analysis to predict potential human target genes. The second objective was to evaluate the role of candidate edible plant-derived miRNAs in the modulation of inflammation-related human genes in human THP-1 macrophages. The third objective was to determine if candidate edible plant-derived miRNAs with potential anti-inflammatory effects could reach the gastrointestinal tract, being detected in human feces and serum samples.
2 Materials and methods
2.1 Total RNA isolation from samples of edible plant-derived foods
Plant fruits (apples, oranges, and pears), vegetables and greens (green peppers, spinaches, green beans, lettuces, and tomatoes), fats and oils (walnuts and olives), legumes (chickpeas and lentils), and cereals (rice) were purchased at local supermarkets. Plant-origin products were considered as different food types or groups: fruits (n = 3), vegetables/greens (n = 5), fats and oils (n = 2), and cooked legumes/cereals (n = 3). Green beans were heat treated in boiling water for 4.5 min in a pressure cooker. Legumes were soaked in water (lentils for 2 h and chickpeas for 12 h) and cooked in a pressure cooker (lentils for 15 min and chickpeas for 30 min). Rice was cooked in water for 20 min in a casserole. Other food elements were used as raw products (non-heat treated).
Total RNA was isolated using the miRNeasy Serum/Plasma Kit (Qiagen, Hilden, Germany) as follows: 0.1 g of fruits, legumes, vegetables, and greens, 0.05–0.1 g of fats and oils (0.05 g of walnut and 0.1 g of olive), and 0.2 g of cereals were added to 1 ml of QIAzol Lysis Reagent and grounded at the highest speed, on ice, using Ultra-Turrax T25 Basic (Basic IKA-Werke, Staufen, Germany) between 1 min (fruits, fats and oils, vegetables, and greens) and 2 min (legumes and cereals). Samples were centrifuged at 12,000 g between 5 min (fruits, walnuts, legumes, and cereals) and 15 min (olives, vegetables, and greens) at 4°C to remove cell debris and insoluble material. The upper-fat layer of olive samples was removed, and olive samples were centrifuged again at 12,000 g for 5 min at 4°C. The supernatants were collected, and RNA purification was concluded following the manufacturer's instructions. For the plant-based food samples used for quantitative real-time PCR (qPCR) assays, 1 μl of spike-in controls (RNA Spike-In Kit, for RT, Qiagen) was added to each sample. RNA for NGS or qPCR was isolated from one sample per type of plant-based food product.
2.2 Next-generation sequencing of miRNAs derived from samples of edible plant-based foods and bioinformatic analysis
NGS was performed in total RNA derived from plant-based products, which were selected for their suitability for small RNA-sequencing (small RNA-seq) after conducting quality controls: 0.1 g of fruits (apple, orange, and pear), 0.1 g of vegetables (spinach and tomato), and 0.05–0.1 g of fats and oil nuts (0.05 g of walnut and 0.1 g of olive). One sample was used for each type of plant-based food product. Libraries were prepared using the NEBNext® Small RNA Library Prep Set for Illumina® kit (New England Biolabs, Ipswich, MA, USA) according to the manufacturer's protocol. Briefly, RNAs were subjected to 3′ and 5′ adaptor ligation and first-strand cDNA synthesis. PCR selectively enriched those DNA fragments that had adapter molecules on both ends. Library amplification was performed by PCR using NEBNext® Multiplex Oligos for Illumina (Index Primers Set 1–4) (New England Biolabs). Purification after PCR was performed using AgenCourt AMPure XP beads (Beckman Coulter, Brea, CA, USA), and libraries were analyzed using Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) to estimate the quantity and check size distribution. The samples underwent bead-based purification to remove big fragments: the samples were incubated with AgenCourt AMPure XP beads at a ratio of 1.3×, and the beads were then discarded. New beads were added to the supernatant for a final ratio of 3.7×, and all material left was recovered. Final libraries were quantified by qPCR using the KAPA Library Quantification Kit (KAPA Biosystems Inc., Wilmington, MA, USA) before amplification with Illumina's cBot. Libraries were sequenced 1 × 50 + 8 bp on Illumina's HiSeq2500.
The bioinformatic analyses to identify miRNA sequences were carried out as follows: the adapter from raw data was removed using a skewer (31), and reads with a size ranging from 15 to 30 bases were aligned to a plant-based reference genome (Prunus persica, NCBIv2, and annotation NCBIv2.52 restricted to miRNAs) (hhttps://plants.ensembl.org/Prunus_persica/Info/Index; https://mirbase.org/results/?query=prunus+persica), with miRNA annotation from miRBase (version 22) using ShortStack (32), allowing a maximum of two mismatches in the seed area. Mapped tags were counted using Htseq-count (33), considering the strand information. Raw reads were normalized with DESeq2. Sequencing data were deposited in NCBI's Gene Expression Omnibus (34): GEO series accession number GSE234786 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE234786).
2.3 Bioinformatic target gene prediction analysis
The RNA target analysis servers psRNATarget scoring schemas V1 and V2 (https://www.zhaolab.org/psRNATarget/) (35) and TAPIR (http://bioinformatics.psb.ugent.be/webtools/tapir/) (36) were used to predict human putative target genes of all the plant-derived miRNAs identified by NGS.
The cDNA library chosen for both psRNATarget and TAPIR servers was the “Homo sapiens (human), transcript, Human genomic sequencing project,” available at psRNATarget, and the bioinformatic predictions were performed with the default parameters. Applying psRNATarget scoring schemas V1 and V2, the following parameters for each output were reported: target accession number of the putative target gene, expectation (penalty for the mismatches), mRNA target aligned fragment, and the inhibitory effect (cleavage or translation inhibition) (35). The scoring Schema V1 also showed the unpaired energy (UPE), which is the energy required to open a mRNA secondary structure (35). The results showed applying the TAPIR program includes the score (penalty for mismatches, gaps, and G-U pairs), the minimum free energy (MFE) ratio, which is a ratio between the free energy of the miRNA–mRNA duplex vs. the free energy of this duplex with perfect matches, and the mRNA target aligned fragment (36). The GeneCodis4 program (https://genecodis.genyo.es/) was used to perform the Gene Ontology (GO) biological process analysis of candidate putative target genes.
2.4 miRNA miR482f and miR482c-5p expression analysis
Expression of miR482f and miR482c-5p was analyzed using qPCR in total RNA isolated from 0.1 g of fruits (apple, orange, and pear), 0.1 g of vegetables and greens (green pepper, spinach, raw and cooked green beans, lettuce, and tomato), 0.05–0.1 g of fats and oils (0.05 g of walnut and 0.1 g of olive), 0.1 g of cooked legumes (chickpeas and lentils), 0.2 g of cooked cereals (rice), biological (serum and feces) human samples (described in MM Section 2.8), and cell culture samples (described in MM Section 2.5). Expression of the spike-in UniSp4 was used as a positive control in plant-derived food, and human samples to determine that short RNA fragments/miRNAs were unaffected by RNA isolation and expression analysis. miR-141-3p and miR-103a-3p were used as human (internal) controls to determine the quality of serum and feces samples and the reliability of the results. Previous evidence has reported consistent presence and stability of miR-141-3p and miR-103a-3p in the feces and serum of mammals, including humans (37–40).
Total RNA (4 μl) was reverse transcribed in 10 μl of final volume reaction with miRCURY LNA RT Kit (Qiagen). Reactions were performed in a MyCycler Thermal Cycler (Bio-Rad, Hercules, CA, USA) at 42°C for 60 min and 95°C for 5 min. cDNA from plant-derived food samples was centrifuged for 1 min at maximum speed, and the supernatant was collected for qPCR.
qPCR was performed with cDNA diluted 1/10 using the miRCURY LNA SYBR Green PCR Kit (Qiagen) and miRCURY LNA miRNA PCR Assays (Qiagen). The sequences of the PCR assays are described in Table 1. qPCR was performed in a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) with the following cycling conditions: 95°C for 2 min, 40 cycles at 95°C for 10 s, and 56°C for 1 min. Non-template control samples were added to each qPCR reaction. qPCR duplicates were made for each sample.
Table 1.
Assays used for qPCR miRNA expression analyses.
| miRNA name | miRBase accession number | Assay reference (GenGlobe ID) | Assay sequence (5′-3′) |
|---|---|---|---|
| hsa-miR-141-3p | MIMAT0000432 | YP00204504 | 5′-UAACACUGUCUGGUAAAGAUGG-3′ |
| hsa-miR-103a-3p | MIMAT0000101 | YP00204063 | 5′-AGCAGCAUUGUACAGGGCUAUGA-3′ |
| ppe-miR482f | MIMAT0031512 | YP02105458 | 5′-UCUUUCCUACUCCACCCAUUCC-3′ |
| ppe-miR482c-5p | MIMAT0027283 | YP02114485 | 5′GGAAUGGGCUGUUUGGGAUG-3′ |
| UniSp4 | N/A | YP00203953 | N/A |
MiRNA names, accession number, and sequences of miRCURY LNA miRNA PCR Assays (Qiagen). N/A, non-applicable.
2.5 Cell culture and edible plant-derived miRNA mimic transfection
Human monocytic leukemia cell line THP-1 was purchased from the American Type Culture Collection (ATCC® TIB-202™; Manassas, VA, USA). THP-1 cells were maintained in RPMI-1640 medium (Gibco, Thermo Fisher Scientific Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin–streptomycin solution (P/S; Gibco) and incubated at 37°C and 5% CO2.
For the experiments, THP-1 cells were seeded in 12-well plates (200,000 cells/well) for gene and miRNA expression assays and 48-well plates (50,000 cells/well) for cytotoxicity assays. The cells were seeded in RPMI-140 medium supplemented with 10% FBS, 1% P/S, and phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, San Luis, MO, USA) at a final concentration of 50 ng/ml to induce the differentiation of THP-1 monocytes into macrophages. The cells were incubated for 48 h.
After differentiation, the cells were forward transfected with Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific Inc., Waltham, MA, USA) and 60 nM of mirVana™ miRNA mimics (Thermo Fisher Scientific Inc.): miR482f (5′-UCUUUCCUACUCCACCCAUUCC-3′), miR482c-5p (5′-GGAAUGGGCUGUUUGGGAUG-3′), and a scramble control (mirVana™ miRNA mimic, negative control #1). Cell medium was changed to RPMI-140 supplemented with 10% FBS, without antibiotics (1 ml/well of 12-well plates, 250 μl/well of 48-well plates). MiRNA mimics and Lipofectamine were diluted in Opti-MEM I Reduced Serum Medium (Gibco), mixed in a 1:1 ratio, and incubated for 5 min at room temperature. miRNA mimic–Lipofectamine complexes were added to each well (100 μl/well of 12-well plates and 25 μl/well of 48-well plates). The volumes of Lipofectamine used were: 2.5 μl per well of 12-well plates (on a volume of 1,100 μl) and 0.625 μl per well of 48-well plates (on a volume of 275 μl). The cells were incubated for 6 h to evaluate the transfection efficiency (detection of transfected miRNAs in cell cultures) or 48 h for gene expression and cytotoxicity assays.
2.6 Gene expression analysis
Cells were frozen on dry ice and stored at −80°C. For miRNA expression analysis experiments, before freezing, the cells were washed with PBS. Total RNA was isolated with TRIzol Reagent (Thermo Fisher Scientific Inc.), according to the manufacturer's instructions. RNA quantity and purity (260/280 ratio) were determined in a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc.). miRNA miR482f and miR482c-5p expression analyses were performed as described in MM Section 2.4.
For mRNA expression analysis, RNA (1 μg) was treated with DNA-free™ DNA Removal Kit (Invitrogen. Thermo Fisher Scientific Inc.) following the manufacturer's protocol. DNA-free RNA was reverse transcribed into cDNA with dNTP Mix (Bioline, Luckenwalde, Germany), Random Primers (Invitrogen. Thermo Fisher Scientific Inc.), Recombinant RNAsin Ribonuclease Inhibitor (Promega, Madison, WI, USA), and M-MLV Reverse Transcriptase (Invitrogen). All the reactions were performed in a GeneAmp PCR System 2700 (Applied Biosystems. Thermo Fisher Scientific Inc.). qPCR was performed with cDNA diluted 1/1.5 using iTaq™ Universal Probes Supermix (Bio-Rad) and specific TaqMan® Gene Expression Assays (Thermo Fisher Scientific Inc.) and Predesigned qPCR Assays from Integrated DNA Technologies (Coralville, IA, USA) described in Table 2. Amplification reactions were performed in a CFX384 Touch Real-Time PCR Detection System (Bio-Rad) as follows: 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min. Non-template control samples were added to each qPCR reaction. Gene expression levels (Cq) were normalized (ΔCq) with the housekeeping gene TATA box binding protein (TBP). The 2−ΔΔCq method (41) was used to determine the relative gene expression of genes, through comparisons between the plant-derived miRNA mimic treatments and the scramble control. At least, two independent experiments were conducted, and qPCR was run in triplicates for each sample.
Table 2.
Assays used for qPCR gene expression analyses.
| Gene name | Assay ID | Accession number (RefSeq) |
|---|---|---|
| NFAM1 | 1Hs.PT.58.45296389 | NM_145912(1) |
| CLEC7A | 1Hs.PT.58.3686547.g | NM_022570(6) |
| TLR6 | 1Hs.PT.58.26737272.g | NM_006068(1) |
| BCL2L12 | 1Hs.PT.58.4708679 | NM_001040668(2) |
| IL10 | 2Hs00961622_m1 | NM_000572.2 |
| TNF | 2Hs00174128_m1 | NM_000594.3 |
| IL1B | 2Hs01555410_m1 |
NM_000576.2 XM_017003988.1 |
| TBP | 2Hs00427620_m1 |
NM_001172085.1 NM_003194.4 |
Gene names, assay IDs, and accession numbers of Integrated DNA technologies Predesigned qPCR Assays1 and TaqMan® Gene Expression Assays2.
2.7 Cytotoxicity assays
Cell viability and proliferation after miRNA treatments were determined with the (3-4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay (CellTiter 96® Aqueous One Solution Cell Proliferation Assay. Promega) in THP-1 macrophages. Cells were incubated with MTS reagent (20 μl of reagent per 100 μl of culture medium) for 2.5 h at 37°C. The absorbance was measured at 490 nm in an absorbance microplate reader Agilent BioTek 800 TS (Agilent Technologies). The cell viability was expressed as the % relative viability; calculated as the mean percentage of each experimental group relative to the mean of the scramble control. Four independent experiments were conducted.
2.8 Detection of edible plant-derived miR482f and miR482c-5p in biological human samples
Plant-derived miRNA analyses from serum and fecal samples were performed as a proof of concept from anonymous volunteers (>18 yr), who qualitatively consumed plant-based products. The samples were donated by nine volunteers (five men and four women) anonymously, and no personal data were collected from these subjects. Specifically, volunteers usually consumed these plant-based food groups: legumes, fruits, vegetables, cereals, and fats and oils, in the variety and proportion they preferred.
Blood samples were collected through a venous puncture in BD Vacutainer® SST™ II Advance tubes (BD Vacutainer Systems, Plymouth, United Kingdom). Samples were kept at room temperature for 30 min and centrifuged at 2,000 g for 15 min at 4°C. Serum was collected and centrifuged at 16,000 g for 10 min at 4°C. Total RNA was extracted from 200 μl of serum samples using miRNeasy Serum/Plasma Advanced Kit (Qiagen). Fecal samples were collected in stool nucleic acid collection and preservation tubes (Norgen Biotek Corp., ON, Canada) according to the manufacturer's instructions. Total RNA was isolated using the RNeasy PowerMicrobiome Kit (Qiagen) from 250 μl of feces following the manufacturer's protocol. Overall, 1 μl of spike-in controls (Qiagen) was added to the serum and fecal samples before RNA extraction. MiRNA miR482f and miR482c-5p expression analyses were performed as described in MM Section 2.4.
2.9 Statistical analysis
The statistical analyses were performed using GraphPad Prism 6.0 for Windows (GraphPad Software Inc., La Jolla, CA, USA). For in vitro studies, comparisons between the miRNA negative control and the experimental groups were performed using Student's t-test. Student's t-test was selected because the aim was to determine differences between plant-derived miR482f and miR482c-5p with the negative control, as independent studies, and not to establish a comparison between the three groups. Statistical significance was considered at p-value <0.05.
3 Results
3.1 Plant-derived miRNAs are present in edible plant-based products, and miR482f and miR482c-5p could potentially inhibit the expression of pro-inflammatory human genes
We performed NGS analysis to identify miRNAs in edible plant-based foods, including fruits (apple, orange, and pear), vegetables (spinach and tomato), and fats and oils (walnut and olive). After the alignment of the reads with the peach genome, as a reference genome, 176 miRNAs were identified in the overall pool of plant-based food samples analyzed (Supplementary Table S1).
The bioinformatic analysis with psRNATarget and TAPIR predicted potential human target genes of the 176 miRNAs detected in the plant-based food samples (data not shown). In the present study, we only focused on miR482f (Ensembl gene: ENSRNA049996209) and miR482c (Ensembl gene ENSRNA049996546) because human gene prediction analyses revealed their potential role in targeting inflammation-related human genes.
The alignment of the miR482f mature sequence with the human transcriptome revealed that 6 (psRNATarget scoring schema V1), 24 (psRNATarget scoring schema V2), and 5 (TAPIR) transcripts could be putative targets (Supplementary Table S2). Altogether, 26 different transcripts (corresponding to 22 different genes) were predicted as putative targets of miR482f. Only three transcripts appeared in the three algorithms. These were as follows: the transcript NM_145912, which codifies for NFAT activating protein with ITAM motif 1 (NFAM1), and the transcripts NM_197949 and NM_022570, which codify for two isoforms of the C-type lectin domain containing 7A (CLEC7A) protein (Supplementary Table S2, Supplementary Figure S1).
The alignment of the plant-derived miR482c-5p mature sequence and the human transcriptome reported that 2 (psRNATarget scoring schema V1), 24 (psRNATarget scoring schema V2), and 5 (TAPIR) transcripts could be putative target genes (Supplementary Table S3). A total of 27 different transcripts (22 different genes) were identified as putative targets of plant-derived miR482c-5p. The only common targets of the three prediction programs were the transcript NM_001160332 of the neurofascin (NFASC) protein and the transcript NM_006068, which encodes for the Toll-like receptor 6 (TLR6) (Supplementary Table S3, Supplementary Figure S2).
According to the bibliography, the combination of psRNATarget and TAPIR programs gives highly accurate predictions (42). Therefore, we selected for subsequent analyses the common target genes to psRNATarget (schemas V1 and V1) and TAPIR: NFAM1 and CLEC7A as putative target genes of miR482f, and NFASC and TLR6 as putative target genes of miR482c-5p.
To explore the biological functions shared between the predicted target genes, we performed Gene Ontology (GO) biological process analysis with GeneCodis4 (Supplementary Figure S3). For the miR482f potential targets, the results revealed that both NFAM1 and CLEC7A participate in the inflammatory response and in the positive regulation of DNA-binding transcription activity (Supplementary Figure S3A). GeneCodis4 associated TLR6 with a wide variety of functions related to the immune system, such as macrophage activation, and pro-inflammatory cytokines production (Supplementary Figure S3B). We did not delve into the study of the potential role of miR482c-5p in shaping the NFASC gene expression since the aim of the present study was to evaluate the impact of plant-derived miRNAs on inflammation. NFASC main functions are related to the nervous system (43). According to Protein Atlas (https://www.proteinatlas.org/ENSG00000163531-NFASC/subcellular), its expression is negligible in THP-1 monocytes, which was the in vitro model selected in this study to achieve the abovementioned objective.
Regarding miR482f, we identified 220.38 normalized reads in the walnut sample, which were also detected in the tomato, orange, olive, and pear samples (Table 3). miR482c, which comprises−5p and−3p sequences, was mostly detected in the walnut and pear samples (655.13 and 266.26 normalized reads, respectively) as well as in the apple, orange, spinach, and olive samples (Table 3), although in lower copy read numbers.
Table 3.
Detection of miR482f and miR482c in plant-based foods by next-generation sequencing.
| Gene ID | miRNA name | Normalized reads | ||||||
|---|---|---|---|---|---|---|---|---|
| Walnut | Tomato | Orange | Pear | Apple | Olive | Spinach | ||
| gene:ENSRNA049996209 | miR482f | 220.38 | 78.09 | 45.35 | 0.10 | U | 0.41 | U |
| gene:ENSRNA049996546 | miR482c | 655.13 | U | 0.60 | 266.26 | 67.70 | 0.28 | 0.27 |
Small RNA-seq was performed with total RNA isolated from 0.05 to 0.1 g of fats and oils and 0.1 g of fruits and vegetables. Reads were mapped to the Prunus persica genome (annotation NCBIv2.52). Results correspond to the normalized reads of a single sample per plant-based product. U, undetectable.
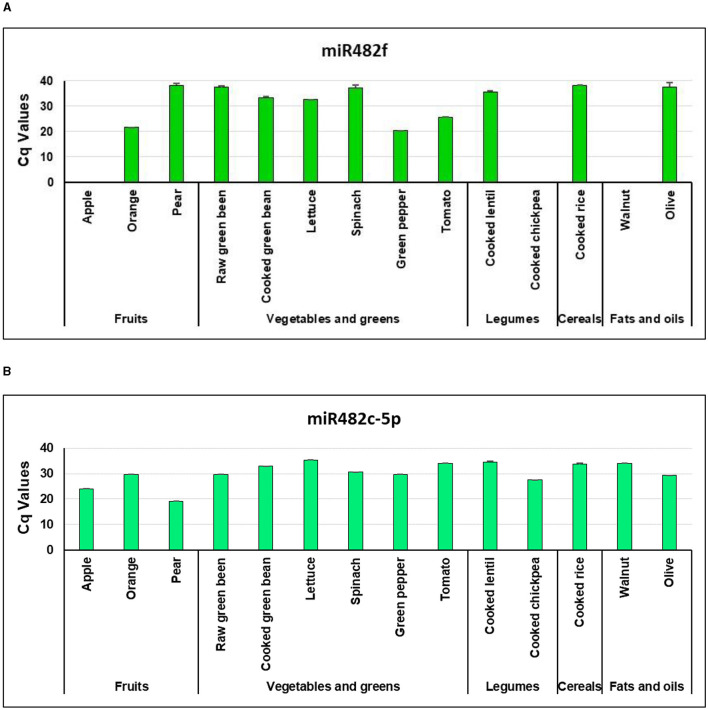
We next sought to qualitatively complement the small RNA-seq results using qPCR analysis in different food matrices, including a greater range of edible plant-derived foods: fruits (apples, oranges, and pears), vegetables and greens (green pepper, spinach, raw and cooked green beans, lettuce, and tomato), fats and oils (walnuts and olives), cooked legumes (chickpeas and lentils), and cooked cereals (rice) (Figure 1). miR482f was detected in fruits (orange and pear), vegetables and greens (raw and cooked green beans, lettuce, spinach, green pepper, and tomato), legumes (cooked lentils), cereals (cooked rice), and fats and oils (olive) (Figure 1A). Regarding miR482c, we validated that the sequence corresponding to the transcript miR482c-5p. miR482c-5p was present in all the selected edible plant-derived foods (Figure 1B). The objective of this analysis was to determine the presence or absence of miRNAs, rather than comparing their abundance between samples. Therefore, data were not normalized to any small RNA sequence (housekeeping) as usually performed for qPCR methods, providing that (1) our aim required only a qualitative measurement and (2) no standardized normalization method has been established for plant-derived miRNA quantification. Positive detection of the spike-in UniSp4 was achieved in all food-based samples, which confirmed the accurate isolation and detection of miRNAs (data not shown).
Figure 1.
Detection of miR482f and miR482c-5p in plant-based foods by qPCR. Cq values of (A) miR482f (5′-UCUUUCCUACUCCACCCAUUCC-3′) and (B) miR482c-5p (5′-GGAAUGGGCUGUUUGGGAUG-3′) with RNA isolated from 0.1 g of fruits (apple, orange, and pear), 0.1 g of vegetables and greens (green pepper, spinach, raw and cooked green beans, lettuce, and tomato), 0.05–0.1 g of fats and oils (0.05 g of walnut and 0.1 g of olive), 0.1 g of cooked legumes (chickpea and lentil), and 0.2 g of cooked cereals (rice). Results are the mean ± standard error of the mean (SEM) (one sample per type of food product and duplicates for qPCR).
Overall, miR482f and miR482c-5p were present in the different raw and cooked plant-derived foods that were analyzed. The miRNA target gene prediction algorithms and the Genecodis4 GO biological processes analyses, suggest that miR482f and miR482c-5p could potentially inhibit the expression of genes involved in inflammation. Therefore, we delved into the study of the potential role of miR482f and miR482c-5p modulating the expression of their putative target genes (NFAM1 and CLEC7A, and TLR6, respectively), and their potential impact on the expression profile of anti- and pro-inflammatory cytokines, in an in vitro model of human macrophages.
3.2 Plant-derived miR482f and miR482c-5p mimics are present in human macrophage-like THP-1 cells after transfection
Human monocytes THP-1 cells differentiated to macrophages were transfected with miRNA mimics miR482f and miR482c-5p, and a scramble control at a concentration of 60 nM. To determine the effectiveness of transfections, we performed qPCR analysis to detect the miRNAs after 6 h of transfection (Supplementary Figure S4). miR482f and miR482c-5p were present in THP-1 cells transfected with each of these mimics (Cq values between 12 and 13). The plant-derived miRNA sequences were not detected in negative control-transfected cells, suggesting that miR482f and miR482c-5p have no human homologous miRNAs in the THP-1 cells.
3.3 Impact of plant-derived miR482f and miR482c-5p mimics on macrophage-like THP-1 viability and/or proliferation
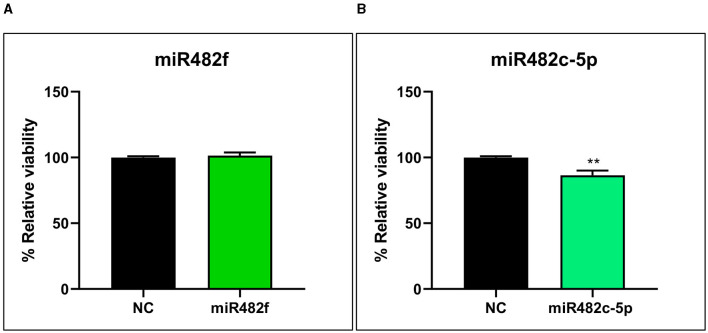
The viability/proliferation of THP-1 macrophage-like cells was evaluated with MTS at 48 h of transfection with 60 nM of plant-derived miR482f, miR482c-5p mimics, and a scramble sequence as a control (Figure 2). miR482f mimic did not induce any change in cell viability or proliferation (Figure 2A), whereas miR482c-5p mimic decreased 13.5 ± 3.6% of the viability/proliferation of THP-1-like macrophages in comparison with the control cells (Figure 2B).
Figure 2.
Evaluation of the effect of plant-derived miRNA mimics (A) miR482f and (B) miR482c-5p on the viability/proliferation of THP-1 macrophage-like cells. THP-1 monocytes differentiated to macrophages for 48 h were transfected with 60 nM of miRVana mimics miR482f (5′-UCUUUCCUACUCCACCCAUUCC-3′), miR482c-5p (5′-GGAAUGGGCUGUUUGGGAUG-3′), and a scramble sequence as a negative control (NC). Cell viability/proliferation was evaluated in THP-1 macrophage-like cells after 48 h of transfections. Results are the mean percentage ± standard error of the mean (SEM) relative to the cells transfected with the scramble sequence (negative control) (n = 4 independent experiments; NC: n = 13; miR482f: n = 13; miR482c-5p: n = 13). NC, negative control. Significance refers to the effect of miR482f or miR482c-5p with respect to the negative control. p-value: ** p < 0.01.
3.4 Plant-derived miR482f and miR482c-5p modulate gene expression of predicted target genes and inflammation-related biomarkers in THP-1 cells
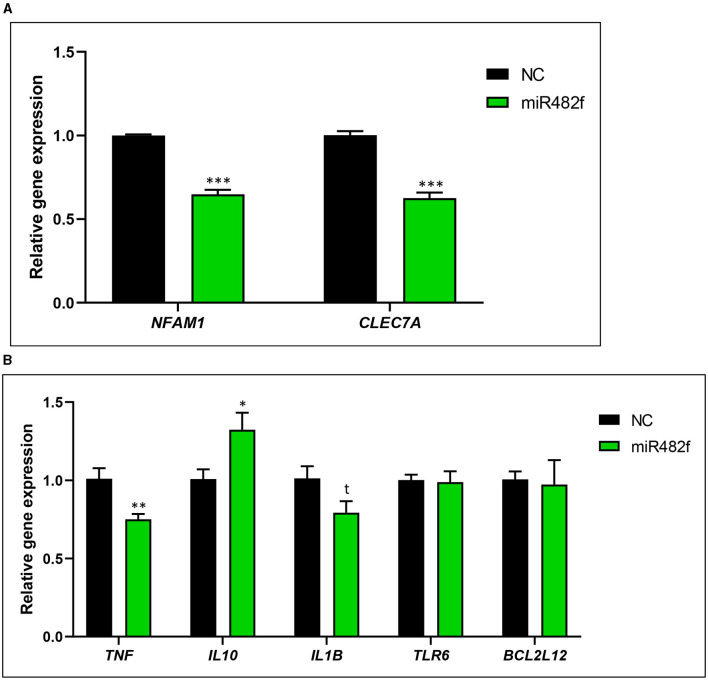
We validated the human target gene bioinformatic predictions (NFAM1 and CLEC7A for miR482f; and TLR6 for mi482c-5p) in the transfected THP-1 macrophage-like cells. Gene expression profile of anti- and pro-inflammatory biomarkers (interleukin 10, IL10; tumor necrosis factor, TNF; interleukin 1 Beta; and IL1B) and the Bcl-2-like protein 12 (BCL2L12) was evaluated.
miR482f mimic downregulated the gene expression of the putative targets NFAM1 (35.2% ± 3.0; p < 0.0001) and CLEC7A (37.7% ± 4.2; p < 0.0001) (Figure 3A) after 48 h of miRNA transfection. miR482f mimic increased the mRNA levels of the anti-inflammatory cytokine IL10 (31.5% ± 13.3, p < 0.05) and reduced the mRNA levels of the pro-inflammatory cytokine TNF (26.0% ± 7.2, p < 0.01) (Figure 3B). No significant changes were detected for IL1B, TRL6, and BCL2L12 gene expression (Figure 3B). A trend was observed for IL1B mRNA decrease (21.9 % ± 10.7, p = 0.0708).
Figure 3.
Gene expression analysis of THP-1 macrophage-like cells transfected with plant-derived miR482f mimic. (A) mRNA levels of putative targets NFAM1 and CLEC7A. (B) Gene expression levels (mRNA) of pro-inflammatory (TNF, IL1B, and TLR6) and anti-inflammatory (IL10) biomarkers and BCL2L12. THP-1 macrophage-like cells were transfected with 60 nM of miRVana mimic miR482f (5′-UCUUUCCUACUCCACCCAUUCC-3′) and a scramble sequence as a negative control. Gene expression levels were measured at 48 h of transfection by qPCR. Results are the gene expression levels of each gene normalized to the housekeeping gene TBP and calculated by the 2−ΔΔCt method, establishing comparisons with the negative control. NC, negative control. The results are presented as mean percentage ± standard error of the mean (SEM) (n = 2 independent experiments, run in triplicate; NC: n = 5; miR482f: n = 6). p-value: * p < 0.05, ** p < 0.01, *** p < 0.001, t = 0.0708.
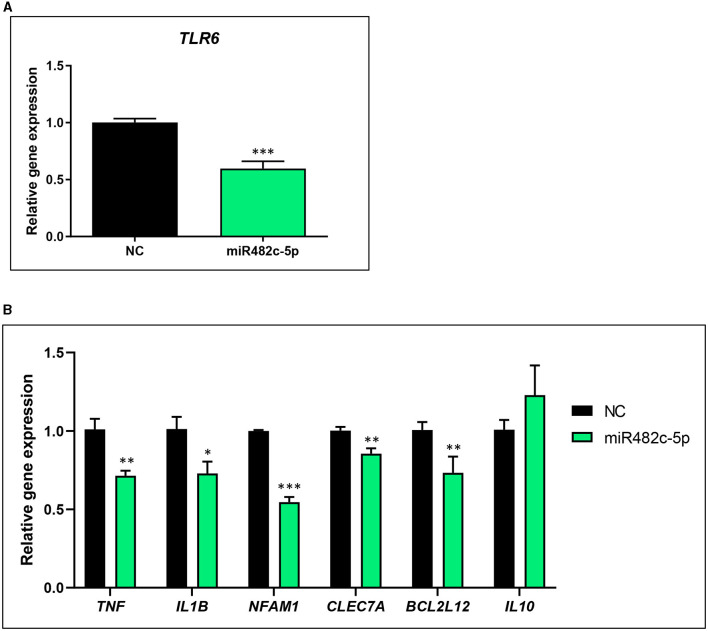
The transfection of miR482c-5p mimic reduced the mRNA levels of the putative target gene TLR6 (40.5% ± 7.6 %, p < 0.001) at 48 h of transfection (Figure 4A). miR482c-5p downregulated gene expression of the pro-inflammatory genes TNF (29.6% ± 7.5, p < 0.01), NFAM1 (45.4% ± 3.4, p < 0.0001), CLEC7A (14.7% ± 4.4 p < 0.01), IL1B (28.4% ± 10.93 p < 0.05), and BCL2L12 (27.39% ± 8.2, p < 0.01), but it did not induce significant changes in the expression of the anti-inflammatory cytokine IL10 (Figure 4B).
Figure 4.
Gene expression analysis of THP-1 macrophage-like cells transfected with plant-derived miR482c-5p mimic. (A) mRNA levels of putative target TLR6. (B) Gene expression levels (mRNA) of pro-inflammatory (TNF, IL1B, TLR6) and anti-inflammatory (IL10) biomarkers and BCL2L12. THP-1 macrophage-like cells were transfected with 60 nM of miRVana mimic miR482c-5p (5′-GGAAUGGGCUGUUUGGGAUG-3′) and a scramble sequence as a negative control. Gene expression levels were measured at 48 h of transfection by qPCR. Results are the gene expression levels of each gene normalized to the housekeeping gene TBP and calculated by the 2−ΔΔCt method, establishing comparisons with the negative control. NC, negative control. The results are presented as mean percentage ± standard error of the mean (SEM) (n = 2 independent experiments, run in triplicate; NC: n = 5; miR482f: NFAM1, TNF, n = 5; CLEC7A, BCL2L12, IL10, IL1B, n = 6). p-value: * p < 0.05, ** p < 0.01, *** p < 0.001. Plant-derived miRNAs miR482f and miR482c-5p are detected in human feces but not in serum.
3.5 Plant-derived miRNAs miR482f and miR482c-5p are detected in human feces but not in serum
To determine whether miR482f and miR482c-5p derived from plant-based foods could reach the gastrointestinal tract, resist degradation during digestion, and potentially be absorbed, we collected feces and serum samples from healthy volunteers who usually consume edible plant-based products. The positive expression of the spike-in UniSp4 in both serum and feces samples confirmed the accuracy of the experimental procedure (RNA isolation and expression analysis) (data not shown).
We measured expression levels of plant-derived miR482f and miR482c-5p using qPCR in nine fecal samples (Figure 5). Both miR482f and miR482c-5p were detected in all the samples, suggesting that plant-derived miRNAs are present in the gastrointestinal tract and resist digestion. The expression of the human hsa-miR-141-3p was used as an endogenous control to determine the quality of the fecal samples.
Figure 5.
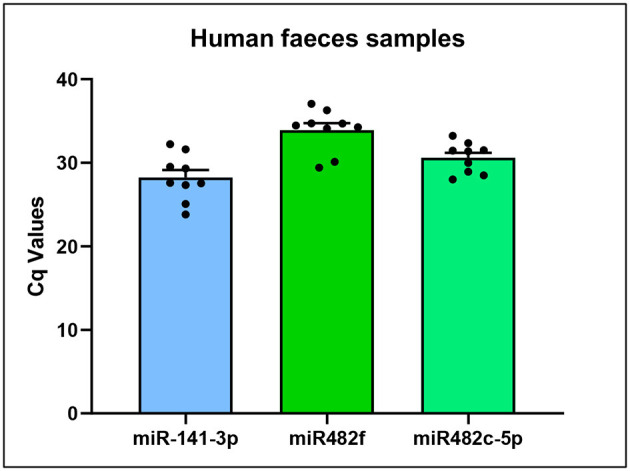
Detection of plant-derived miR482f and miR482c-5p in human feces of healthy volunteers using qPCR. Cq values of plant-derived miR482f (5′-UCUUUCCUACUCCACCCAUUCC-3′), miR482c-5p (5′-GGAAUGGGCUGUUUGGGAUG-3′), and human miR-141-3p (5′-UAACACUGUCUGGUAAAGAUGG-3′) as an endogenous control, in nine human feces samples (qPCR duplicates for each sample) from healthy volunteers. Results are the mean ± standard error of the mean (SEM) (n = 9).
We analyzed the levels of plant-derived miR482f and miR482c-5p in serum samples from the same volunteers in which the two plant-derived miRNAs were present in feces. Neither miR482f nor miR482c-5p were detected in serum samples in any of the volunteers (data not shown). To further confirm the reliability of the results, we used the human miR-103a-3p as an endogenous control. hsa-miR-103a-3p was detected in all the serum samples analyzed (Cq values between 24 and 28), suggesting that the lack of detection of plant-derived miR482f and miR482c-5p could be due to their very low or inexistent quantity to be detected by qPCR and not because of the serum sample quality.
4 Discussion
The present study aimed to identify miRNAs from edible plant-derived foods that could modulate the expression of human genes associated with inflammation. The rationale of this hypothesis is mainly based on the evidence supporting (1) the role of plant-based diets in alleviating inflammation, and (2) the recent discovery of miRNAs as bioactive molecules of plants and cross-kingdom gene expression regulators (19, 44).
We selected five groups of plant-derived foods (fruits, vegetables and greens, legumes, cereals, and fats and oils) based on previous evidence that unveiled their anti-inflammatory properties (8–11). Although we identified a wide variety of miRNAs in edible plants, miR482f and miR482c-5p were selected for their potential role in targeting inflammatory human genes. We found that these miRNAs are present in the five groups of foods. We used the Prunus Persica genome as a reference to annotate plant-derived miRNA sequences of edible plants detected by NGS. As regards our results, both NGS and qPCR, there is a high degree of conservation of these miRNAs in the plant kingdom or at least among the plant-origin food products analyzed in this study. The results revealed that original plant-derived miR482f and miR482c withstand standard heat-treatment (cooking) processes. In agreement with our findings, other authors have also reported that harsh conditions such as storage, processing, and cooking do not compromise plant-derived miRNA survival (45). For some plant-based food samples, we detected miR482f or miR482c by NGS and not by qPCR and vice versa. For instance, we detected miR482f in walnuts only by NGS and in spinach only by qPCR. The miR482c were detected in tomato only by qPCR. The results obtained with both techniques are complementary, given that the nature of the sample might determine the suitability for either NGS and/or qPCR. However, the NGS hits that could not be validated by qPCR should be taken with caution. Reliable conclusions can only be drawn about the presence or absence of miR482f and miR482c in each plant-based product (verified by two independent methods). The relative abundance between different types of plant-based products cannot be compared since there is no reliable and universal endogenous plant-derived miRNA normalization method established yet. Our in silico analysis predicted that our miRNA candidates (miR482f and miR482c-5p) would eventually bind specific mRNA targets associated with inflammation. Providing that the mechanism of action of miRNA usually links to mRNA inhibition (target repression), plant-derived miR482f and miR482c-5p could eventually downregulate inflammation-related genes. To eventually validate this hypothesis, we used differentiated human THP-1 macrophages, an in vitro cell model widely used in the study of the antioxidant and anti-inflammatory bioactive properties of chemical compounds and derived food products (46–48).
Using differentiated human THP-1 macrophages, we have demonstrated for the first time that plant-derived miR482f and miR482c-5p can achieve efficient inhibition of their predicted targets (CLEC7A and NFAM1 and TLR6, respectively) and induce significant changes in other (functional) inflammation-related genes. As regards our results, plant-derived miR482f and miR482c-5p stand out as bioactive molecules from dietary sources with the potential to manage inflammation-associated genes. However, specific differences (in direct targets and in functional genes) between both miRNAs exist and could suggest different but complementary mechanisms of action for each plant-derived miRNA. miR482f and miR482c-5p direct targets had previously been linked to immune system function (49–52) and production of pro-inflammatory cytokines (such as TNF-α, IL1-β, and IL-6) (53–56), suggesting that their inhibition could have a therapeutic approach. The finding that miR482c-5p also reciprocally downregulated CLEC7A and NFAM1 gene expression as its counterpart (miR482f) is of relevance since a positive correlation between TLR6 and CLEC7A expression has also been reported (57). On the other hand, while miR482f reciprocally upregulated anti-inflammatory (IL10) and downregulated pro-inflammatory factors (TNF), miR482c-5p only downregulated pro-inflammatory genes (TNF, IL1B). Whether these changes at the mRNA expression level translate to functional (i.e., protein secretion) modifications remains to be established in future studies.
Interestingly, it has been documented that TLRs could modulate the expression of anti-apoptotic Bcl-2 family proteins, and TLR6 could protect cells from apoptosis (58–61). This evidence could explain the slight decrease in the viability/proliferation that we detected upon the inhibition of TLR6 by miR482c-5p. In concordance, the downregulation of the anti-apoptotic gene BCL2L12 (62, 63), in miR482c-5p treated cells suggests that this miRNA could be interfering with the viability of macrophages. We hypothesize that, since in miR482c-5p-treated cells the TLR6 levels decrease, these cells are less protected against pro-apoptotic stimuli than other (scramble control and miR482f-treated) cells in which the expression of TLR6 was not affected. We speculate that TLR6 inhibition might not decrease cell viability in physiological conditions absent of potentially harmful events (i.e., transfection procedure). In any case, we consider that the decrease in macrophage viability (10%) is very low to be considered biologically relevant.
The role of plant-derived miRNAs as cross-kingdom immune-modulatory agents has recently been documented. This could be mediated through plant-derived miRNA interaction with viral, bacteria, and mammalian cell genes (64–68). In the present study, we reported a direct interaction between plant-derived miRNAs with immunomodulatory properties and mammalian genes, which has been also documented by other studies (67, 68). Indeed, we found that plant-derived miRNAs could interact with immune system cells, in particular with human macrophages, and regulate gene expression. Although the mechanism underlying the interactions between plant-derived miRNAs and immune system cells described in our study relies mainly on gene expression regulation, we do not exclude the involvement of other additional/complementary mechanisms (i.e., involving indirect target gene interactions) (68).
A crucial but controversial aspect of the plant-derived miRNA cross-kingdom gene expression regulation hypothesis is the bioavailability in the host. Interestingly, edible plant-derived miRNAs could be present in extracellular vesicles (EVs) (69, 70), which could be a key determinant of absorption (16, 71, 72). Nevertheless, bioactive activities upon the absorption by mammalian cells have been reported in vivo after the administration of plant-derived miRNAs in a free form (68, 73). This evidence suggests that either in a free form or encapsulated in EVs, plant-derived miRNAs could have a cross-kingdom functional impact. In the present study, we verified that plant-derived miR482f and miR482c-5p were detected in human feces but not in the serum of healthy individuals who usually consumed plant-based products. These results demonstrate that plant-derived miRNAs (miR482f and miR482c-5p) resist physical (cooking and digestive motility) and chemical treatments (digestive enzymes) but is rather unlikely that they could reach physiological levels in the bloodstream. We do not rule out completely the possibility that miR482f and miR482c-5p could be absorbed since it is quite reasonable that their bioavailability might be lower (or beyond that) the detection level of standard methods and conditions used for addressing small RNA expression. It could be worthy to analyze their presence in serum in other experimental conditions since the bioavailability of miRNAs might depend on dietary intake levels (74), gut permeability variability (16), and the sensitivity of the techniques used for its detection. Interestingly, it has been suggested that technical limitations, artifacts, contaminations, inter-individual variability in gut permeability, and dietary abundance and stability of plant-derived miRNAs could explain the disparity of the results reported between studies (75, 76). Importantly, plant-derived miRNAs could potentially modulate inflammation at the local (gut) level through the modulation of tissue-resident immune cells (macrophages, lymphocytes) and/or gut microbiota. Although some previous evidence supports this hypothesis (66), intimate mechanisms of this cross-kingdom communication remain to be established; especially, the direct interaction of plant-derived miRNAs with gut immune cells should be confirmed in future in vivo and/or clinical studies. The abundance of plant-derived miR482f and miR482c-5p in fecal samples suggests that they reach the gastrointestinal tract (and move forward throughout the intestinal lumen). As regards our findings, plant-derived miRNAs are physically present and stable in the gastrointestinal tract. Whether their presence in host organisms fully translates into physiological changes (at cellular/tissue level) remains to be established in future experiments since it would require additional resources (methodologies, ethical) or invasive procedures (biopsies) and this was out of our scope. Although in vivo approaches would eventually contribute to clarify this, important inter-species differences arise. For instance, in silico plant-derived miR482f and miR482c-5p predictions are biologically different in human vs. mouse (according to their respective mRNA sequence) and thus prevent the implementation of animal models in the context of our study.
This article is not free of limitations. Additional research would be necessary to explore the role of plant-derived miR482f and miR482c-5p on inflammation beyond the regulation of gene expression levels. To determine if plant-derived miR482f and miR482c-5p effects at the gene level could translate to physiological events such as cytokine protein expression changes, we evaluated IL-10 secretion levels in conditioned media, as a first preliminary (proof of concept) approach. We found that these miRNAs modulated IL-10 secretion levels (data not shown), concordantly to reported gene expression changes. However, depth investigations will be required to evaluate the overall impact of plant-derived miR482f and miR482c-5p on the protein secretion levels of the whole cytokines panel presented in this study (a secretome analysis). Thus, whether the modulation of inflammatory-associated genes promoted by plant-derived miR482f and miR482c-5p fully translate to physiologically relevant events remains to be established in future studies. This could determine whether these miRNAs might potentially be important therapeutic agents to shape immune system responses.
5 Conclusion
We show here that miR482f and miR482c-5p are present in standard plant-based foods (legumes, vegetables, greens, fruits, cereals, and fats and oils), resist physical–chemical degradation during cooking and digestion, and reach the human gut, although eventually, they are not absorbed. In vitro, these plant-derived miRNAs modulate the gene expression profile of inflammation-associated biomarkers in human macrophages. A better understanding of the cross-talk between plant-derived miR482f and miR482c-5p and the immune system could help to determine their therapeutic potential as bioactive molecules with anti-inflammatory properties.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE234786.
Ethics statement
The studies involving humans were approved by University of Navarra, Pamplona, Spain. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
ED-S: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. SL-C: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing. PA: Methodology, Writing – review & editing. E-ZA: Writing – review & editing. JR-B: Funding acquisition, Project administration, Writing – review & editing. FM: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Acknowledgments
The authors thank the Centre for Genomic Regulation (CRG) for performing the next-generation sequencing of miRNAs derived from plant-based samples and bioinformatic analyses to identify plant-derived miRNA sequences.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Spanish Biomedical Research Centre in Physiopathology of Obesity and Nutrition (CIBERobn, grant No. CB12/03/30002), Ministerio de Asuntos Económicos y Transformación Digital (PID2022-141766OB-I00, RTI2018-102205-B-I00, and BFU2015-65937-R projects), and a Center for Nutrition Research pre-doctoral grant awarded to ED-S.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2023.1287312/full#supplementary-material
References
- 1.Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, et al. Chronic inflammation in the etiology of disease across the life span. Nat Med. (2019) 25:1822–32. 10.1038/s41591-019-0675-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajendran P, Chen YF, Chen YF, Chung LC, Tamilselvi S, Shen CY, et al. The multifaceted link between inflammation and human diseases. J Cell Physiol. (2018) 233:6458–71. 10.1002/jcp.26479 [DOI] [PubMed] [Google Scholar]
- 3.Zhao H, Shang Q, Pan Z, Bai Y, Li Z, Zhang H, et al. Exosomes from adipose-derived stem cells attenuate adipose inflammation and obesity through polarizing m2 macrophages and beiging in white adipose tissue. Diabetes. (2018) 67:235–47. 10.2337/db17-0356 [DOI] [PubMed] [Google Scholar]
- 4.Rohm TV, Meier DT, Olefsky JM, Donath MY. Inflammation in obesity, diabetes, and related disorders. Immunity. (2022) 55:31–55. 10.1016/j.immuni.2021.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng J, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. (2018) 9:7204–18. 10.18632/oncotarget.23208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grosso G, Laudisio D, Frias-Toral E, Barrea L, Muscogiuri G, Savastano S, et al. Anti-inflammatory nutrients and obesity-associated metabolic-inflammation: state of the art and future direction. Nutrients. (2022) 14:1137. 10.3390/nu14061137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichelmann F, Schwingshackl L, Fedirko V, Aleksandrova K. Effect of plant-based diets on obesity-related inflammatory profiles: a systematic review and meta-analysis of intervention trials. Obes Rev. (2016) 17:1067–79. 10.1111/obr.12439 [DOI] [PubMed] [Google Scholar]
- 8.Xiao Y, Xia J, Ke Y, Cheng J, Yuan J, Wu S, et al. Effects of nut consumption on selected inflammatory markers: a systematic review and meta-analysis of randomized controlled trials. Nutrition. (2018) 54:129–43. 10.1016/j.nut.2018.02.017 [DOI] [PubMed] [Google Scholar]
- 9.Hosseini B, Berthon BS, Saedisomeolia A, Starkey MR, Collison A, Wark PAB, et al. Effects of fruit and vegetable consumption on inflammatory biomarkers and immune cell populations: a systematic literature review and meta-analysis. Am J Clin Nutr. (2018) 108:136–55. 10.1093/ajcn/nqy082 [DOI] [PubMed] [Google Scholar]
- 10.Juárez-Chairez MF, Meza-Márquez OG, Márquez-Flores YK, Jiménez-Martínez C. Potential anti-inflammatory effects of legumes: a review. Br J Nutr. (2022) 128:2158–69. 10.1017/S0007114522000137 [DOI] [PubMed] [Google Scholar]
- 11.Milesi G, Rangan A, Grafenauer S. Whole grain consumption and inflammatory markers: a systematic literature review of randomized control trials. Nutrients. (2022) 14:374. 10.3390/nu14020374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majdan M, Bobrowska-Korczak B. Active compounds in fruits and inflammation in the body. Nutrients. (2022) 14:2496. 10.3390/nu14122496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajaram S, Damasceno NR, Braga RA, Martinez R, Kris-Etherton P, Sala-Vila A. Effect of nuts on markers of inflammation and oxidative stress: a narrative review. Nutrients. (2023) 15:1099. 10.3390/nu15051099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Awika JM, Rose DJ, Simsek S. Complementary effects of cereal and pulse polyphenols and dietary fiber on chronic inflammation and gut health. Food Funct. (2018) 9:1389–409. 10.1039/C7FO02011B [DOI] [PubMed] [Google Scholar]
- 15.Li D, Yang J, Yang Y, Liu J, Li H, Li R, et al. A timely review of cross-kingdom regulation of plant-derived MicroRNAs. Front Genet. (2021) 12:613197. 10.3389/fgene.2021.613197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Díez-Sainz E, Lorente-Cebrián S, Aranaz P, Riezu-Boj JI, Martínez JA, Milagro FI. Potential mechanisms linking food-derived microRNAs, gut microbiota and intestinal barrier functions in the context of nutrition and human health. Front Nutr. (2021) 8:586564. 10.3389/fnut.2021.586564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. Metazoan MicroRNAs. Cell. (2018) 173:20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Q, Hu B, Zhang C. MicroRNAs and their roles in plant development. Front Plant Sci. (2022) 13:824240. 10.3389/fpls.2022.824240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Xu R, Li N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr Metab (Lond). (2018) 15:68. 10.1186/s12986-018-0305-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saiyed AN, Vasavada AR, Johar SRK. Recent trends in miRNA therapeutics and the application of plant miRNA for prevention and treatment of human diseases. Futur J Pharm Sci. (2022) 8:24. 10.1186/s43094-022-00413-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T, Ma F, Peng Y, Sun R, Xi Q, Sun J, et al. Plant miR167e-5p promotes 3T3-L1 adipocyte adipogenesis by targeting β-catenin. Vitr Cell Dev Biol Anim. (2022) 58:471–9. 10.1007/s11626-022-00702-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. (2012) 22:107–26. 10.1038/cr.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Chen T, He J-J, Wu J-H, Luo J-Y, Ye R-S, et al. Plant MIR167e-5p inhibits enterocyte proliferation by targeting β-Catenin. Cells. (2019) 8:1385. 10.3390/cells8111385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akao Y, Kuranaga Y, Heishima K, Sugito N, Morikawa K, Ito Y, et al. Plant hvu-MIR168-3p enhances expression of glucose transporter 1 (SLC2A1) in human cells by silencing genes related to mitochondrial electron transport chain complex I. J Nutr Biochem. (2022) 101:108922. 10.1016/j.jnutbio.2021.108922 [DOI] [PubMed] [Google Scholar]
- 25.Marzano F, Caratozzolo MF, Consiglio A, Licciulli F, Liuni S, Sbisà E, et al. Plant miRNAs reduce cancer cell proliferation by targeting MALAT1 and NEAT1: a beneficial cross-kingdom interaction. Front Genet. (2020) 11:552490. 10.3389/fgene.2020.552490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu J, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. (2015) 25:39–49. 10.1038/cr.2014.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minutolo A, Potestà M, Gismondi A, Pirrò S, Cirilli M, Gattabria F, et al. Olea europaea small RNA with functional homology to human miR34a in cross-kingdom interaction of anti-tumoral response. Sci Rep. (2018) 8:12413. 10.1038/s41598-018-30718-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundaram GM. Dietary non-coding RNAs from plants: Fairy tale or treasure? Non-coding RNA Res. (2019) 4:63–8. 10.1016/j.ncrna.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang F, Du J, Liang Z, Xu Z, Xu J, Zhao Y, et al. Large-scale analysis of small RNAs derived from traditional Chinese herbs in human tissues. Sci China Life Sci. (2019) 62:321–32. 10.1007/s11427-018-9323-5 [DOI] [PubMed] [Google Scholar]
- 30.Huang H, Davis CD, Wang TTY. Extensive degradation and low bioavailability of orally consumed corn miRNAs in mice. Nutrients. (2018) 10:215. 10.3390/nu10020215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang H, Lei R, Ding S-W, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. (2014) 15:182. 10.1186/1471-2105-15-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Axtell MJ. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. (2013) 19:740–51. 10.1261/rna.035279.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. (2015) 31:166–9. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. (2002) 30:207–10. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. (2011) 39:W155–9. 10.1093/nar/gkr319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonnet E, He Y, Billiau K, Van de Peer Y. TAPIR a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. (2010) 26:1566–8. 10.1093/bioinformatics/btq233 [DOI] [PubMed] [Google Scholar]
- 37.Moloney GM, Viola MF, Hoban AE, Dinan TG, Cryan JF. Faecal microRNAs: indicators of imbalance at the host-microbe interface? Benef Microbes. (2018) 9:175–83. 10.3920/BM2017.0013 [DOI] [PubMed] [Google Scholar]
- 38.Wohnhaas CT, Schmid R, Rolser M, Kaaru E, Langgartner D, Rieber K, et al. Fecal MicroRNAs show promise as noninvasive crohn's disease biomarkers. Crohn's Colitis. (2020) 2:otaa003. 10.1093/crocol/otaa003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Bian Q-Z, Zhang W, Cui H-B. Circulating microRNA-103a-3p could be a diagnostic and prognostic biomarker for breast cancer. Oncol Lett. (2022) 23:38. 10.3892/ol.2021.13156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wanhainen A, Mani K, Vorkapic E, De Basso R, Björck M, Länne T, et al. Screening of circulating microRNA biomarkers for prevalence of abdominal aortic aneurysm and aneurysm growth. Atherosclerosis. (2017) 256:82–8. 10.1016/j.atherosclerosis.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 41.Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2∧(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. (2013) 3:71–85. [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava PK, Moturu TR, Pandey P, Baldwin IT, Pandey SP. A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genomics. (2014) 15:348. 10.1186/1471-2164-15-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zonta B, Tait S, Melrose S, Anderson H, Harroch S, Higginson J, et al. Glial and neuronal isoforms of Neurofascin have distinct roles in the assembly of nodes of Ranvier in the central nervous system. J Cell Biol. (2008) 181:1169–77. 10.1083/jcb.200712154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giugliano D, Ceriello A, Esposito K. The effects of diet on inflammation: emphasis on the metabolic syndrome. J Am Coll Cardiol. (2006) 48:677–85. 10.1016/j.jacc.2006.03.052 [DOI] [PubMed] [Google Scholar]
- 45.Philip A, Ferro VA, Tate RJ. Determination of the potential bioavailability of plant microRNAs using a simulated human digestion process. Mol Nutr Food Res. (2015) 59:1962–72. 10.1002/mnfr.201500137 [DOI] [PubMed] [Google Scholar]
- 46.Chanput W, Mes JJ, Wichers HJ. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol. (2014) 23:37–45. 10.1016/j.intimp.2014.08.002 [DOI] [PubMed] [Google Scholar]
- 47.Siegel PM, Sander L, Fricke A, Stamm J, Wang X, Sharma P, et al. P(2)Y(12) receptor blockers are anti-inflammatory drugs inhibiting both circulating monocytes and macrophages including THP-1 cells. Sci Rep. (2021) 11:17459. 10.1038/s41598-021-95710-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Nasser MM, Al-Dosari MS, Parvez MK, Al-Anazi MR, Alkahtane AA, Alothaid H, et al. The potential effects of Indigofera coerulea extract on THP-1 human cell line. J King Saud Univ - Sci. (2021) 33:101446. 10.1016/j.jksus.2021.101446 [DOI] [Google Scholar]
- 49.Juchem KW, Gounder AP, Gao JP, Seccareccia E, Yeddula N, Huffmaster NJ, et al. NFAM1 promotes pro-inflammatory cytokine production in mouse and human monocytes. Front Immunol. (2021) 12:773445. 10.3389/fimmu.2021.773445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang C, Kamiya T, Liu Y, Kadoki M, Kakuta S, Oshima K, et al. Inhibition of dectin-1 signaling ameliorates colitis by inducing lactobacillus-mediated regulatory T cell expansion in the intestine. Cell Host Microbe. (2015) 18:183–97. 10.1016/j.chom.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 51.Pedro AR V Lima T Fróis-Martins R Leal B Ramos IC Martins EG . Dectin-1-mediated production of pro-inflammatory cytokines induced by yeast β-glucans in bovine monocytes. Front Immunol. (2021) 12:689879. 10.3389/fimmu.2021.689879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castoldi A, Andrade-Oliveira V, Aguiar CF, Amano MT, Lee J, Miyagi MT, et al. Dectin-1 activation exacerbates obesity and insulin resistance in the absence of MyD88. Cell Rep. (2017) 19:2272–88. 10.1016/j.celrep.2017.05.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. (2000) 97:13766–71. 10.1073/pnas.250476497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joosten LAB, Abdollahi-Roodsaz S, Dinarello CA, O'Neill L, Netea MG. Toll-like receptors and chronic inflammation in rheumatic diseases: new developments. Nat Rev Rheumatol. (2016) 12:344–57. 10.1038/nrrheum.2016.61 [DOI] [PubMed] [Google Scholar]
- 55.Zou M, Yang W, Niu L, Sun Y, Luo R, Wang Y, et al. Polydatin attenuates Mycoplasma gallisepticum (HS strain)-induced inflammation injury via inhibiting the TLR6/MyD88/NF-κB pathway. Microb Pathog. (2020) 149:104552. 10.1016/j.micpath.2020.104552 [DOI] [PubMed] [Google Scholar]
- 56.Arias-Loste MT, Iruzubieta P, Puente Á, Ramos D, Santa Cruz C, Estébanez Á, et al. Increased expression profile and functionality of TLR6 in peripheral blood mononuclear cells and hepatocytes of morbidly obese patients with non-alcoholic fatty liver disease. Int J Mol Sci. (2016) 17:1878. 10.3390/ijms17111878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moreira AP, Cavassani KA, Ismailoglu UB, Hullinger R, Dunleavy MP, Knight DA, et al. The protective role of TLR6 in a mouse model of asthma is mediated by IL-23 and IL-17A. J Clin Invest. (2011) 121:4420–32. 10.1172/JCI44999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abrahams VM, Aldo PB, Murphy SP, Visintin I, Koga K, Wilson G, et al. TLR6 modulates first trimester trophoblast responses to peptidoglycan. J Immunol. (2008) 180:6035–43. 10.4049/jimmunol.180.9.6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garg M, Potter JA, Abrahams VM. Identification of microRNAs that regulate TLR2-mediated trophoblast apoptosis and inhibition of IL-6 mRNA. PLoS ONE. (2013) 8:e77249. 10.1371/journal.pone.0077249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jablonska E, Garley M. TLRs and Bcl-2 family proteins in neutrophils of oral cavity cancer patients. Folia Histochem Cytobiol. (2009) 47:615–9. 10.2478/v10042-008-0118-8 [DOI] [PubMed] [Google Scholar]
- 61.Parker LC, Whyte MKB, Dower SK, Sabroe I. The expression and roles of Toll-like receptors in the biology of the human neutrophil. J Leukoc Biol. (2005) 77:886–92. 10.1189/jlb.1104636 [DOI] [PubMed] [Google Scholar]
- 62.Stegh AH, Kim H, Bachoo RM, Forloney KL, Zhang J, Schulze H, et al. Bcl2L12 inhibits post-mitochondrial apoptosis signaling in glioblastoma. Genes Dev. (2007) 21:98–111. 10.1101/gad.1480007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li M-G, Liu X-Y, Liu Z-Q, Hong J-Y, Liu J-Q, Zhou C-J, et al. Bcl2L12 contributes to Th2-biased inflammation in the intestinal mucosa by regulating CD4(+) T cell activities. J Immunol. (2018) 201:725–33. 10.4049/jimmunol.1800139 [DOI] [PubMed] [Google Scholar]
- 64.Teng Y, Xu F, Zhang X, Mu J, Sayed M, Hu X, et al. Plant-derived exosomal microRNAs inhibit lung inflammation induced by exosomes SARS-CoV-2 Nsp12. Mol Ther. (2021) 29:2424–40. 10.1016/j.ymthe.2021.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minutolo A, Potestà M, Roglia V, Cirilli M, Iacovelli F, Cerva C, et al. Plant microRNAs from moringa oleifera regulate immune response and HIV infection. Front Pharmacol. (2020) 11:620038. 10.3389/fphar.2020.620038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, et al. Plant-derived exosomal micrornas shape the gut microbiota. Cell Host Microbe. (2018) 24:637–652.e8. 10.1016/j.chom.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou D, He F, Ma L, Cao M, Zhou Z, Wei Z, et al. The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J Nutr Biochem. (2018) 57:197–205. 10.1016/j.jnutbio.2018.03.026 [DOI] [PubMed] [Google Scholar]
- 68.Cavalieri D, Rizzetto L, Tocci N, Rivero D, Asquini E, Si-Ammour A, et al. Plant microRNAs as novel immunomodulatory agents. Sci Rep. (2016) 6:25761. 10.1038/srep25761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao J, Feng S, Wang X, Long K, Luo Y, Wang Y, et al. Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. PeerJ. (2018) 6:e5186. 10.7717/peerj.5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baldrich P, Rutter BD, Karimi HZ, Podicheti R, Meyers BC, Innes RW. Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-Nucleotide “Tiny” RNAs. Plant Cell. (2019) 31:315–24. 10.1105/tpc.18.00872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Del Pozo-Acebo L, López de Las Hazas M-C, Margollés A, Dávalos A, García-Ruiz A. Eating microRNAs: pharmacological opportunities for cross-kingdom regulation and implications in host gene and gut microbiota modulation. Br J Pharmacol. (2021) 178:2218–45. 10.1111/bph.15421 [DOI] [PubMed] [Google Scholar]
- 72.Urzì O, Gasparro R, Ganji NR, Alessandro R, Raimondo S. Plant-RNA in extracellular vesicles: the secret of cross-kingdom communication. Membranes. (2022) 12:352. 10.3390/membranes12040352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li M, Chen T, Wang R, Luo J-Y, He J-J, Ye R-S, et al. Plant MIR156 regulates intestinal growth in mammals by targeting the Wnt/β-catenin pathway. Am J Physiol Cell Physiol. (2019) 317:C434–48. 10.1152/ajpcell.00030.2019 [DOI] [PubMed] [Google Scholar]
- 74.Yang J, Farmer LM, Agyekum AAA, Elbaz-Younes I, Hirschi KD. Detection of an abundant plant-based small RNA in healthy consumers. PLoS ONE. (2015) 10:e0137516. 10.1371/journal.pone.0137516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kang W, Bang-Berthelsen CH, Holm A, Houben AJS, Müller AH, Thymann T, et al. Survey of 800+ data sets from human tissue and body fluid reveals xenomiRs are likely artifacts. RNA. (2017) 23:433–45. 10.1261/rna.059725.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Witwer KW. Alternative miRNAs? Human sequences misidentified as plant miRNAs in plant studies and in human plasma. F1000Research. (2018) 7:244. 10.12688/f1000research.14060.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE234786.