Abstract

Neuroinflammation plays a vital role in Alzheimer’s disease (AD) pathogenesis and other neurodegenerative disorders (NDs). Presently, only symptomatic treatments are available and no disease-modifying drugs are available for AD and other NDs. Thus, targeting AD-associated neuroinflammation with anti-inflammatory compounds and antioxidants has recently been given much focus. Now, flavonoids are being increasingly investigated as therapeutic agents to treat inflammation; apigenin has a neuroprotective effect. Iron dyshomeostasis plays a key role in sustaining the neuroinflammatory phenotype, highlighting the importance of maintaining iron balance, in which human transferrin (HTF) plays a vital role in this aspect. Herein, we explored the binding and dynamics of the HTF–apigenin complex using multifaceted computational and experimental approaches. Molecular docking revealed that apigenin occupies the iron-binding pocket of HTF, forming hydrogen bonds with critical residues Arg475 and Thr686. Molecular dynamics simulations deciphered a dynamic view of the HTF–apigenin complex’s behavior (300 ns) and suggested that the complex maintained a relatively stable conformation. The results of spectroscopic observations delineated significant binding of apigenin with HTF and stable HTF–apigenin complex formation. The observed binding mechanism and conformational stability could pave the way for developing novel therapeutic strategies to target neuroinflammation by apigenin in the context of iron homeostasis.
1. Introduction
In the present times, neurodegenerative disorders (NDs) pose a serious threat to public health. No available treatments to cure NDs, such as Parkinson’s disease (PD), and Alzheimer’s disease (AD), are among the main reasons. However, there is an increasing range of therapeutic and supportive options available.1 AD is a ND that involves progressive loss of neurons in the brain2 and is among the most common forms of dementia currently affecting 44 million people worldwide, and expect this number to increase exponentially in the near future.3 We do not know the precise etiology for most AD patients suffering from sporadic or late-onset AD. Still, aging and inheritance of the ε4 allele of the apolipoprotein E gene are vital risk factors.4 Amyloid β (Aβ) is a pathological hallmark protein of AD that aggregates, resulting in the formation of extracellular plaques. These plaques affect various cellular functions,5 ranging from alterations of the regulatory checks of calcium signaling systems to the generation of reactive oxygen (ROS) and nitrogen species (RNS).6 Moreover, Aβ accumulation also results in inflammation that leads to microglia activation around plaques.7 Neurofibrillary tangles, the second major pathological AD hallmarks, are formed due to hyperphosphorylation of tau protein that ultimately destabilizes microtubules.8 As per recent literature, there is increasing evidence to decipher neuroinflammation as a major pathological component of AD.9 In many recent reports, a strong correlation is established between AD pathogenesis and neuroinflammation; according to a recent study, brain’s inflammatory response contributed primarily in AD pathogenesis.10,11 According to a study by Hesse et al., the brains of AD patients showed high levels of pro-inflammatory mediators.12 Astrocytes and microglia are the cells that comprise the central nervous system (CNS), and they play a vital role in physiology and disease. Glial activation is a defensive mechanism that controls tissue repair in the initial stages of neurodegeneration.13 Though unnecessary and extended activation leads to a chronic neuroinflammatory response,14 that is implicated in most of NDs onset and progression, viz. amyotrophic lateral sclerosis, PD, and AD.15−18 Due to the activation of microglia and astrocytes, inflammatory mediators are produced, namely, cytokines, chemokines, ROS, RNS, and others that create an environment of neuroinflammation ultimately resulting in neuronal death.
All of these reports highlight the importance of neuroinflammation in AD pathogenesis. For many years, intensive global research has explored new domains in the context of NDs therapeutics to identify and develop new drug treatments. In spite of these intensifying efforts, to date, only symptomatic treatments are available and no disease-modifying drugs are available for AD and other NDs. Thus, targeting AD-associated neuroinflammation with anti-inflammatory compounds and antioxidants is attracting researchers’ attention and much focus is given to it in present times.19−21
Studies have documented that AD progression and onset can be reduced successfully by dietary-based neuroprotective, anti-inflammatory, and antioxidant agents.22 Flavonoids are naturally occurring phenolic compounds that are increasingly being explored as therapeutic agents to treat inflammation, owing to their powerful effects in controlling inflammation. Many pieces of literature document the association of flavonoid consumption resulting in reduced dementia levels, strengthening the importance of these flavonoids as anti-inflammatory agents.23,24 Flavonoids, a diverse class of natural compounds, are widely present in edible plants, vegetables, fruits, and various plant-based food items; the fundamental structures consist of three interconnected cyclic carbon rings. Specifically, two of these rings are benzene rings designated as A and B rings, and they are joined via a heterocyclic pyran ring denoted as the C ring. Flavonoids can be categorized into subclasses, including flavonols, flavones, flavanones, anthocyanidins, isoflavones, dihydroflavonols, and chalcones. These classifications are based on factors such as the linkage positions of the B and C rings, as well as the extent of saturation, oxidation, and hydroxylation of the C ring.25 Apigenin, structurally 4′,5,7-trihydroxyflavone, is a flavonoid that belongs to a class of flavones and is abundantly present in teas and fruits.26,27 It can effectively cross the blood–brain barrier, making it a vital cog in treating CNS disorders, and is also nontoxic even at very high doses. The importance of apigenin is attributable to the wide range of biological activities associated with apigenin, ranging from antioxidant28 to neuroprotective6 and others.29,30 According to a recent study, apigenin presents neuroprotective and anti-inflammatory effects in vitro.7 Another study showed that the treatment of interferon gamma-activated microglia with apigenin led to the reduced production of pro-inflammatory IL-6 and TNFα through mechanisms involving STAT1.31 According to another report, apigenin protected Aβ25–35-induced toxicity in rat cerebral microvascular endothelial cells.32 In another literature report, apigenin treatment of mice enhanced spatial learning and memory after amnesia induction with Aβ25–35.33 All these reports highlight the importance of apigenin as an anti-inflammatory agent and as a neuroprotectant. Neuroinflammation is comorbid with brain iron accumulation, which is apparent in many NDs, viz., AD, PD, and others.
Free iron is an effective neurotoxin that can generate ROS, resulting in oxidative stress.34 Iron, in its free form, is a potent neurotoxin that can induce oxidative stress, which is a key pathological feature of neurodegeneration. Oxidative stress is created so that highly reactive radicals, viz. hydroxyl radicals are generated via a Fenton reaction that can ultimately damage DNA, proteins, and lipids, resulting in cell death, a key pathological feature of NDs. The generation of ROS and RNS, which are directly involved in the inflammatory process, can significantly affect iron metabolism via their interaction with iron-regulatory proteins.
In NDs, the stimulus for neuroinflammation is unclear, but after activation of microglia and astrocytes, there is a chronic and sustained response. Iron dyshomeostasis is also apparent, playing a key role in sustaining the neuroinflammatory phenotype. Human transferrin (HTF) is a key player in iron homeostasis and ferritin. HTF, one of the foremost serum proteins in plasma,35 plays a vital role as a Fe transporter; it associates with a complex of iron, transferrin receptor and cells’ endosomal compartment.36 The pro-inflammatory cytokines will induce changes in the iron proteins responsible for maintaining iron homeostasis in a way that increased amounts of iron will be deposited in brain cells. Thus, iron loading in specific brain regions in AD arises due to an initial neuro-inflammatory event. Hence, releasing a number of cytokines, ROS and RNS from activated glial cells can modify the activity of iron proteins in maintaining iron homeostasis. Recently, many researchers have given attention to the interaction between HTF and various drugs, viz. AD drugs,37 anticancer drugs,38 and others. A study by Du et al. investigated the structural effects on the conformational transition of HTF induced by the binding of flavonoids with different numbers and positions of hydroxyl groups.65 In a recent investigation, researchers delved into understanding the interaction mechanism between transferrin and four closely related flavonols—galangin, kaempferol, quercetin, and myricetin—utilizing a combination of experimental and computational modeling methods.25 All of the reports signify the importance of iron homeostasis in AD-associated neuroinflammation, thereby revealing the vital role of HTF in AD therapeutics. Thus, in our present work, we will be elucidating the binding and interaction mechanism of apigenin with HTF employing a combined in silico and in vitro approaches.
2. Materials and Methods
2.1. Chemicals and Reagents
HTF and apigenin were procured from Sigma-Aldrich Co. LLC. (St. Louis, MO, USA). We dialyzed HTF overnight in 20 mM sodium phosphate buffer, pH 7.2, to get rid of any excessive salts and checked the purity on SDS-PAGE. We used all other chemicals of analytical grade. In all spectroscopic measurements, we used controls, and the reported spectra are the subtracted spectra. All the experiments were performed in triplicates.
2.2. UV–Vis Spectroscopic Measurements
Absorption spectra of HTF in the presence and absence of apigenin were measured on a Shimadzu UV-1900 Spectrophotometer with a quartz cuvette (1 cm path length). We kept the concentration of Tf constant at 2 μM and varied the apigenin concentration (0–9 μM). The data were fitted into eq 1 as per earlier literature to obtain the binding constant (K) of HTF–apigenin complex.39
| 1 |
2.3. Fluorescence-Based Binding Assay at Varying Temperatures
In triplicate, fluorescence binding was performed on a Shimadzu Spectrofluorometer (RF-6000) (Japan). We titrated HTF with various concentrations of apigenin, and fluorescence emission was recorded in a range between 300 and 400 nm wavelengths with a 280 nm excitation light. To 500 μL of HTF, apigenin was successively added from its stock solution such that its concentration was 0–3.5 μM. We then used the obtained data and put it into the Stern–Volmer (SV) and modified Stern–Volmer (MSV) eqs (eq 2) and analyzed as per earlier published studies37,40 to obtain binding parameters for the HTF–apigenin complex.
| 2 |
F0: fluorescence intensity of protein; F: fluorescence intensity of protein in the presence of ligands; K: binding constant; n: number of binding sites; and C: ligand molecules’ concentration.
2.4. Molecular Docking
The molecular docking analysis was performed by using a DELL workstation running the Windows 11 operating system. The three-dimensional structure of HTF was sourced from the Protein Data Bank (PDB ID: 3V83), while the structure of apigenin was retrieved from the PubChem database (PubChem CID: 5280443) and processed utilizing MGL tools.41 For docking simulations and to unveil intricate ligand–receptor interactions, the InstaDock software was employed.42 The docking procedure was executed using a blind search space approach, encompassing the entire protein and affording ample room for each ligand to explore its potential binding sites. The grid dimensions were set at 82, 99, and 84 Å for the X, Y, and Z dimensions, respectively. The grid center was positioned at coordinates −52.355, 17.601, and −30.21 for the X, Y, and Z axes. A grid spacing of 1 Å was maintained, and default docking parameters were applied. Subsequently, the docking results were visualized and analyzed using Discovery Studio Visualizer43 for figure rendering and comprehensive assessment of the docking outcomes.
2.5. MD Simulations
After the docking investigation, comprehensive all-atom molecular dynamics (MD) simulations were undertaken on HTF and its complex with apigenin. These simulations were carried out over a duration of 300 ns (ns) using the charmm36-jul2022 force field within the GROMACS 2020 beta platform.44 To establish the system for simulations, the ligand topology was generated utilizing the CHARMM General Force Field (CGenFF) program and cgenff_charmm2gmx_py3_nx2.py, followed by its integration with the parent coordinates of HTF. The resulting systems were then solvated in a cubic box with dimensions of 10 Å, utilizing the SPC216 water model through the gmx solvate module.45 Subsequently, an energy minimization step was carried out, employing the steepest descent algorithm with 1500 steps. The trajectories acquired from the MD simulations were analyzed using the GROMACS tools suite. Graphical representations were generated utilizing XMGrace software.46 For a more comprehensive and in-depth insight into the details of the molecular docking and MD simulations, we direct interested readers to our recent publications, where a detailed exposition of the methodologies can be found.47,48
3. Results
3.1. Molecular Docking
The molecular docking studies revealed remarkable interactions between apigenin and HTF. By analyzing the docking results, we were able to identify the interacting residues and investigate the binding pattern of apigenin with HTF. The binding affinity of the HTF–apigenin complex was determined to be −7.8 kcal/mol. Apigenin shows a remarkable ligand efficiency value, i.e., 0.39 kcal/mol/non-H atom, for HTF.
To gain insights into the inhibitory mechanism of apigenin toward HTF, we conducted a detailed analysis of the docking results. Our findings indicated that apigenin preferred to occupy the iron-binding pocket of HTF. Within this deep cavity, apigenin formed significant interactions with the binding pocket residues, as depicted in Figure 1. Notably, one hydrogen bond was observed between apigenin and Thr686, a key residue in the iron-binding site (Figure 1B). Surface representations further illustrated the occupancy of the iron-binding pocket by apigenin within the internal cavity of the HTF (Figure 1B). Furthermore, pivotal interactions, such as alkyl, π-alkyl, and van der Waals forces, ensued with multiple residues of HTF, holding the intricate structure of the complex (Figure 1C). Previous research has similarly documented hydrogen carbonate and iron binding to HTF through a similar set of amino acids (https://www.uniprot.org/uniprotkb/P02787/entry#function).49 Thus, we hypothesize that apigenin binds to HTF within the binding pocket, impeding the entry of hydrogen carbonate. As a consequence, this interference ultimately impedes the catalytic function of HTF.
Figure 1.
Apigenin binding to HTF. (A) Cartoon representation of docked apigenin interacting with HTF. (B) Surface potential view of the HTF binding pocket occupied by apigenin. (C) 2D plot of interacting residues of HTF with apigenin.
3.2. HTF Dynamics upon Apigenin Binding
Utilizing MD simulations, a robust computational approach, we delved into the intricate internal dynamics, physical movements, and conformational alterations of macromolecules. This technique also facilitated the exploration of their interactions with other molecules within a specified time frame.50 Within this study’s context, we undertook extensive all-atom MD simulations, each spanning 300 ns, for both the unbound HTF and the HTF–apigenin complex. Our primary goal was to scrutinize the conformational transitions, overall stability, and interaction mechanisms between apigenin and HTF.
The association of a small molecule with a protein’s binding pocket often prompts significant conformational shifts in its globular architecture.51 To assess the structural variation and stability of the protein, we employed root-mean-square deviation (RMSD), a widely employed metric. Our observations indicate that apigenin binding imparts HTF stability while inducing only minor alterations in its native conformation (Figure 2A). Nevertheless, initial fluctuations in RMSD surfaced during the initial 50 ns post apigenin binding, potentially attributable to the initial positioning of apigenin within HTF’s binding pocket. Subsequent to this phase, the binding event induced RMSD equilibrium throughout the simulations, underscoring the stability of the HTF–apigenin complex (Figure 2A).
Figure 2.
Structural dynamics of HTF with apigenin in (A) RMSD, (B) RMSF, (C) Rg, and (D) SASA plot. Black and red represent values obtained for free HTF and HTF–apigenin complexes, respectively.
Intrigued by the local structure and inherent flexibility, we quantified all residues’ root-mean-square fluctuation (RMSF), elucidating residual fluctuations within both unbound HTF and HTF upon apigenin binding (Figure 2B). The RMSF profile unveils fluctuations in distinct regions of the HTF structure. Notably, the average RMSF of HTF exhibits a similarity post apigenin binding, manifesting sporadic fluctuations and stabilizations across the simulation period, encompassing the N-terminal to the C-terminal domains. However, an amplified level of residual fluctuations is discernible upon apigenin binding, which is primarily concentrated at the N-terminal domain. This occurrence could be attributed to heightened residual vibrations during the simulation stemming from the presence of apigenin. The amalgamation of our MD simulations and subsequent analyses furnishes valuable insights into the dynamic interplay between apigenin and HTF, highlighting stabilizing effects and subtle structural adjustments within the HTF–apigenin complex.
The radius of gyration (Rg) stands as a crucial structural parameter, serving as a window into a protein’s tertiary volume and overall conformational configuration.52 Its widespread application lies in evaluating a protein’s stability within a biological context. A higher Rg value points to relatively looser protein structure packing. This parameter has been harnessed to discern the influence of a protein’s conformational dynamics in its native environment.53 Through meticulous analysis, the average Rg values for both free HTF and the HTF–apigenin complex were gauged to fall within the 2.90–3.05 nm range. Strikingly, the Rg values exhibited remarkable consistency for HTF, both prior to and following apigenin binding, over the entire simulation duration (Figure 2C). Our in-depth examination of the Rg plot uncovered marginal changes, underscoring the absence of substantial structural rearrangements in HTF’s packing subsequent to apigenin binding. This observation does agree with the earlier deduction, reinforcing the notion that apigenin’s interaction elicits minimal alteration in HTF’s native conformation. In summary, the Rg parameter emerges as a pivotal tool in our investigative arsenal, illuminating HTF’s tertiary characteristics and conformational stability. Its unwavering behavior upon apigenin association fortifies the proposition that apigenin’s impact on HTF’s structure is subtle, reinforcing the protein’s structural integrity.
The solvent accessible surface area (SASA) emerges as a pivotal descriptor, delineating the surface region of a protein that remains directly exposed to the surrounding solvent environment.54 SASA intertwines closely with the Rg parameter and holds the potential to offer profound insights into a protein’s behavior. This parameter unveils the extent to which a protein’s surface interacts with its environment, thereby reflecting its conformational dynamism. Our meticulous analysis encompassed the calculation and temporal plotting of average SASA values for both unbound HTF and the HTF–apigenin complex. These values were derived within the range of 290–320 nm2. Interestingly, a subtle elevation in SASA values was manifested when HTF was engaged with apigenin, as compared to HTF alone (Figure 2D). This minute SASA increment suggests the occurrence of conformational shifts in HTF attributed to apigenin’s binding, resulting from the ligand’s occupancy within the protein’s intramolecular space. This phenomenon attests to the interplay between HTF and apigenin, influencing the protein’s overall surface interaction with its surroundings. In synthesis, our comprehensive analyses coalesce to reinforce the theme of conformational stability within the protein–ligand complex throughout the simulation. The SASA parameter is a vital corroborative element, accentuating the nuanced interplay between HTF and apigenin and shedding light on the complex’s enduring structural integrity.
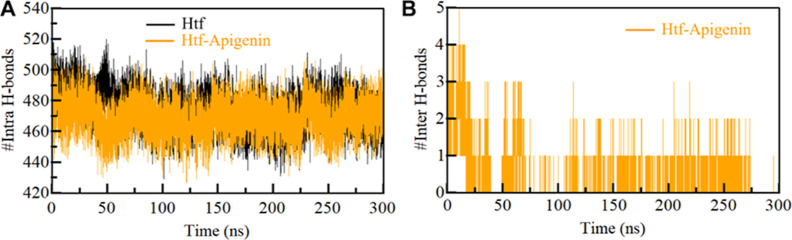
3.3. Hydrogen Bonding Dynamics: Anchoring Protein Stability
The intricate network of intramolecular hydrogen bonding is a basis for maintaining protein stability, conferring both directionality and steadfastness to protein–ligand complexes.55 The exploration of hydrogen bond dynamics affords a unique vantage point, illuminating the stability of the polar interactions within these complexes. Our study undertook a comprehensive analysis, delving into the stability of both unbound HTF and the HTF–apigenin complex by quantifying the number of hydrogen bonds throughout the simulation (Figure 3A). This undertaking provides a compelling depiction of how apigenin’s presence can influence the formation of specific hydrogen bonds within the HTF structure.
Figure 3.
Time evolution of hydrogen bonds formed. (A) Intra-HTF, (B) hydrogen bonds between HTF and apigenin.
Furthermore, we meticulously computed the average number of conventional hydrogen bonds formed between HTF and apigenin. The findings unveiled a dynamic landscape wherein up to 5 hydrogen bonds were established during the simulation period. Notably, these hydrogen bonds exhibited fluctuations of varying intensities, spanning from 2 to 4 bonds and 1 to 2 bonds, characterized by minimal fluctuations. These results mirror our earlier molecular docking outcomes (Figure 3B). These observations serve as a testament to the dynamic formation and fluctuation of hydrogen bonds between HTF and apigenin. These interactions are unequivocally indicative of their engagement within the binding pocket of HTF. This analysis provides a profound glimpse into the intricate interplay of hydrogen bonding, underscoring its pivotal role in modulating the stability of the HTF–apigenin complex. This dynamic interaction enriches our understanding of the complex’s stability and underscores the intricate involvement of hydrogen bonding in mediating protein–ligand interactions.
3.4. Principal Component Analysis
Principal56 component analysis(PCA) is a fundamental method employed to investigate the collective atomic movements within proteins. PCA provides valuable insights into the overall protein dynamics by identifying principal motions characterized by eigenvectors, allowing for exploring its stability and conformational changes.57 In this investigation, we utilized PCA to assess the conformational variability of both free HTF and the HTF–apigenin complex within the essential subspace, as depicted in Figure 4. The tertiary structures projected by the Cα atom along the first and second eigenvectors illustrate the spectrum of conformational states adopted by the protein. A cluster of stable states is evident for HTF, implying that the protein explores a broader range of phase spaces. However, there is no significant global shift in the motion of HTF upon the binding of apigenin (Figure 4B). This indicates that the interaction with apigenin does not trigger substantial large-scale conformational alterations in the protein’s structure.
Figure 4.
PCA (A) 2D projections of HTF conformations. (B) Time evolution conformations of free HTF and HTF–apigenin complexes.
3.5. Free Energy Landscape Analysis
To delve deeper into HTF’s conformational dynamics, we investigated the free energy landscapes (FELs) of HTF and the HTF–apigenin complex, as illustrated in Figure 5. The FEL of HTF reveals a singular, stable global minimum, predominantly confined within a solitary basin (Figure 5A). However, a distinct alteration in the conformational behavior becomes evident upon the introduction of apigenin, resulting in the protein attaining a more extensive energy minimum (Figure 5B). This observation indicates that the binding of apigenin induces a modification in the energy landscape of HTF, leading to the emergence of discrete conformational states. By combining the analysis of conformational dynamics through PCA and FEL exploration, we extract valuable insights into the stability and evolving conformational patterns of HTF, particularly in its interplay with apigenin. These discoveries significantly contribute to our comprehension of the conformational landscape and potential functional implications of HTF in the presence of apigenin.
Figure 5.
FEL for the (A) HTF and (B) HTF–apigenin complex.
3.6. UV Visible Spectroscopy
There are many approaches that can be used to detect complex formations between ligand and protein and UV visible spectroscopy is amongst them.58 Herein, a typical absorption peak of HTF at around 280 nm was observed for the native protein (Figure 6A). In the presence of increasing concentration of apigenin, there was an increase in the absorbance of protein, i.e., hyperchromism was observed, without generating any new peak and no shift in the absorption maxima. It is said that wherein dynamic quenching plays a primary role, no changes in UV visible spectra should be observed owing to the fact that the interaction occurs at the excited state of the fluorophore. Herein, our observations suggest that complex formation is taking place between HTF and apigenin; apigenin binds with HTF, resulting in a stable complex formation. Furthermore, the obtained data were fitted into a double reciprocal plot (Figure 6B) to obtain the binding constant (K) for the HTF–apigenin complex. We obtained a binding constant (K) of 6.5 × 104 M–1 for this complex from the ratio of the intercept and the slope of this plot, implicating a significant strength of binding between apigenin and HTF.
Figure 6.
(A) UV vis absorption spectra of HTF in the absence and presence of apigenin (0–9 μM). (B) Double reciprocal plot of the HTF–apigenin complex.
3.7. Fluorescence Spectroscopy
Fluorescence spectroscopy is a commanding technique to study molecular interactions that involve proteins, attributable to the fact that it is a rapid and simple method apart from being sensitive.59,60 Fluorescence is specifically attributed to the presence of three intrinsic fluorophores in the protein; among them, tryptophan (trp) is the one that contributes maximally.61 HTF’s intrinsic fluorescence is primarily from Trp alone, with phenylalanine having a very low quantum yield. Tyrosine fluorescence is completely quenched if it is ionized or near an amino group, a carboxyl group, or a Trp. Many studies have employed this approach to elucidate the binding mechanism of important ligands/drugs with HTF and other clinically relevant proteins in a way delineating the nature of the binding process.37,40 Intrinsic fluorescence of HTF is very sensitive to its microenvironment62 and thus any changes in the microenvironment around fluorophores due to binding of ligand are reflected in changes in the fluorescence spectra.
HTF, in its native form, exhibited a distinct fluorescence maximum of around 335 nm after excitation at 280 nm. The effect of various apigenin concentrations on the fluorescence intensity of native HTF is shown in Figure 7. It is apparent that apigenin quenched the fluorescence of HTF in a dose-dependent manner with no observed peak shift and/or generation of an extra new peak; i.e., when HTF concentration was kept constant, and apigenin concentration was increased gradually, a visible decrease in the fluorescence intensity of HTF was evident. This phenomenon is termed fluorescence quenching, and this observed loss of HTF’s fluorescence in the presence of apigenin can be attributed to the formation of the HTF–apigenin complex.
Figure 7.

Fluorescence intensity of free HTF and HTF with varying apigenin concentrations (0–3.5 μM). Excitation was performed at 280 nm.
SV and MSV equations were used in line with earlier published studies to compute the different binding parameters for the complex formation.63,64 The SV plot was well-fitted with a linear function (Figure 8A), suggestive of the fact that interaction between HTF and apigenin was predominantly driven by a single mechanism, i.e., either static or dynamic. For this interaction, Ksv found from the slope of the SV plot was 1.38 × 105 M–1 s–1. Furthermore, the quenching data of HTF in the presence of varying apigenin concentrations were fitted in the MSV equation to give an MSV plot (Figure 8B), and the intercept of this plot yielded the binding constant for the HTF–apigenin complex. Apigenin was found to bind to HTF with a binding constant (K) of 1.5 × 105 M–1. The obtained values are in the range reported for other protein–ligand complexes,39,63 implying significant interaction strength between apigenin and HTF. These results and UV visible spectroscopy further validated our in silico observations and affirmed the formation of a stable HTF–apigenin complex.
Figure 8.
(A) SV and (B) MSV plot of the HTF–apigenin complex.
4. Discussion
We comprehensively investigated the molecular interactions between HTF and apigenin, employing combined in silico and in vitro approaches. Molecular docking analysis reveals compelling evidence of a strong interaction between apigenin and HTF. The observed binding affinity of the HTF–apigenin complex underscores the favorable energetics of their interaction. Notably, apigenin’s occupation of the iron-binding pocket of HTF and the formation of key hydrogen bonds with critical residues, such as Arg475 and Thr686, offer crucial insights into the binding mechanism. The study’s findings suggest that apigenin’s binding might disrupt the hydrogen carbonate binding, subsequently affecting HTF’s catalytic function.
The MD simulations provide a dynamic view of the HTF–apigenin complex’s behavior over a prolonged period, i.e., 300 ns. The RMSD analysis illustrates that apigenin binding confers stability to HTF, with minor fluctuations observed during the initial stages of the simulation. The RMSF analysis further elucidates the impact of apigenin on the flexibility of specific regions within the HTF, particularly in the N-terminal domain. This increased flexibility is attributed to apigenin’s presence and highlights its influence on HTF’s internal dynamics. The Rg and SASA analyses contribute to understanding the overall conformational stability and surface interactions of the HTF–apigenin complex, emphasizing the subtle nature of the structural alterations induced by apigenin binding. The hydrogen bond dynamics demonstrate the intricate role of hydrogen bonds in stabilizing the protein–ligand complex. The fluctuating pattern of hydrogen bond formation between HTF and apigenin underscores their dynamic interplay, suggesting a delicate balance between stability and flexibility. The PCA and FEL analyses collectively shed light on the conformational space explored by HTF and the HTF–apigenin complex. The absence of significant global conformational shifts upon apigenin binding indicates that the complex maintains a relatively stable conformation while adapting to subtle local changes. Furthermore, we employed in vitro approaches to validate these observations. Formation of a complex between protein and ligand can be observed through corresponding changes in UV–visible absorption spectra, and the results demonstrated significant binding affinity of apigenin with HTF with a binding constant (K) of 6.5 × 104 M–1. Fluorescence spectroscopy is a probing technique that senses changes in fluorophores’ local environment; any change in the local environment around these fluorophores directly corresponds to changes in fluorescence spectra. Thus, the intrinsic fluorescence of proteins demonstrates significant evidence of their structure and dynamics and serves as a commanding technique to study molecular interactions involving proteins and ligands, revealing the binding mechanism. The results revealed that apigenin binds to HTF with an excellent affinity with a binding constant (K) of the order of 105 M–1, forming a stable complex. Thus, these spectroscopic observations exacerbated the in silico observations, affirming the formation of a stable HTF–apigenin complex.
The study’s approach of combining molecular docking and MD simulations with spectroscopic assays provides a comprehensive understanding of the complex’s behavior at static and dynamic levels. The findings presented here contribute valuable insights into the molecular interactions between HTF and apigenin and provide a solid foundation for future research endeavors aimed at harnessing the therapeutic potential of apigenin in targeting HTF-related disorders.
Acknowledgments
M.S.K. acknowledges and extends their appreciation to the Researchers Supporting Project Number (RSP2023R352), King Saud University, Riyadh, Saudi Arabia for funding this study. A.S. is grateful to Ajman University, UAE for supporting this publication.
Data Availability Statement
The information supporting this study is available in this article. All the generated data is presented in the manuscript.
Author Contributions
Conceptualization, Anas Shamsi; data curation, Moyad Shahwan; formal analysis, Anas Shamsi, Moyad Shahwan, and Saleha Anwar; funding acquisition, Anas Shamsi; investigation, Dharmendra Kumar Yadav; methodology, Anas Shamsi and Moyad Shahwan; project administration, Anas Shamsi; resources, Moyad Shahwan and Mohd Khan; software, Anas Shamsi, Dharmendra Kumar Yadav, and Mohd Khan; validation, Saleha Anwar and Mohd Khan; visualization, Dharmendra Kumar Yadav; writing–original draft, Anas Shamsi, Moyad Shahwan, and Saleha Anwar; and writing–review and editing, Dharmendra Kumar Yadav and Mohd Khan.
The authors declare no competing financial interest.
References
- Szigeti K. Overcoming gaps in the treatment of neurodegenerative disease. EBioMedicine 2020, 60, 103088. 10.1016/j.ebiom.2020.103088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C.; Brayne C.; Mayeux R. Epidemiology of Alzheimer disease. Nat. Rev. Neurol. 2011, 7 (3), 137–152. 10.1038/nrneurol.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olajide O. A.; Sarker S. D. Alzheimer’s disease: natural products as inhibitors of neuroinflammation. Inflammopharmacology 2020, 28 (6), 1439–1455. 10.1007/s10787-020-00751-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier J.; Bertrand P.; Kogan S.; Gauthier S.; Davignon J.; Bouthillier D. Apolipoprotein E polymorphism and Alzheimer’s disease. Lancet 1993, 342 (8873), 697–699. 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- De Strooper B.; Iwatsubo T.; Wolfe M. S. Presenilins and γ-secretase: structure, function, and role in Alzheimer disease. Cold Spring Harbor Perspect. Med. 2012, 2 (1), a006304. 10.1101/cshperspect.a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balez R.; Steiner N.; Engel M.; Muñoz S. S.; Lum J. S.; Wu Y.; Wang D.; Vallotton P.; Sachdev P.; O’Connor M.; et al. Neuroprotective effects of apigenin against inflammation, neuronal excitability and apoptosis in an induced pluripotent stem cell model of Alzheimer’s disease. Sci. Rep. 2016, 6 (1), 31450. 10.1038/srep31450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dourado N. S.; Souza C. d. S.; De Almeida M. M. A.; Bispo da Silva A.; Dos Santos B. L.; Silva V. D. A.; De Assis A. M.; da Silva J. S.; Souza D. O.; Costa M. d. F. D.; et al. Neuroimmunomodulatory and neuroprotective effects of the flavonoid apigenin in in vitro models of neuroinflammation associated with Alzheimer’s disease. Front. Aging Neurosci. 2020, 12, 119. 10.3389/fnagi.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-z.; Wu Q.; Smith A.; Grundke-Iqbal I.; Iqbal K. τ is phosphorylated by GSK-3 at several sites found in Alzheimer disease and its biological activity markedly inhibited only after it is prephosphorylated by A-kinase. FEBS Lett. 1998, 436 (1), 28–34. 10.1016/S0014-5793(98)01090-4. [DOI] [PubMed] [Google Scholar]
- Webers A.; Heneka M. T.; Gleeson P. A. The role of innate immune responses and neuroinflammation in amyloid accumulation and progression of Alzheimer’s disease. Immunol. Cell Biol. 2020, 98 (1), 28–41. 10.1111/imcb.12301. [DOI] [PubMed] [Google Scholar]
- Heppner F. L.; Ransohoff R. M.; Becher B. Immune attack: the role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 2015, 16 (6), 358–372. 10.1038/nrn3880. [DOI] [PubMed] [Google Scholar]
- Fu W.-Y.; Wang X.; Ip N. Y. Targeting neuroinflammation as a therapeutic strategy for Alzheimer’s disease: mechanisms, drug candidates, and new opportunities. ACS Chem. Neurosci. 2019, 10 (2), 872–879. 10.1021/acschemneuro.8b00402. [DOI] [PubMed] [Google Scholar]
- Hesse R.; Wahler A.; Gummert P.; Kirschmer S.; Otto M.; Tumani H.; Lewerenz J.; Schnack C.; von Arnim C. A. Decreased IL-8 levels in CSF and serum of AD patients and negative correlation of MMSE and IL-1β. BMC Neurol. 2016, 16, 185. 10.1186/s12883-016-0707-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit W. J. Microglia and neuroprotection: implications for Alzheimer’s disease. Brain Res. Rev. 2005, 48 (2), 234–239. 10.1016/j.brainresrev.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Kraft A. D.; Harry G. J. Features of microglia and neuroinflammation relevant to environmental exposure and neurotoxicity. Int. J. Environ. Res. Public Health 2011, 8 (7), 2980–3018. 10.3390/ijerph8072980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka M. T.; Carson M. J.; Khoury J. E.; Landreth G. E.; Brosseron F.; Feinstein D. L.; Jacobs A. H.; Wyss-Coray T.; Vitorica J.; Ransohoff R. M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14 (4), 388–405. 10.1016/s1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadery C.; Koshimori Y.; Coakeley S.; Harris M.; Rusjan P.; Kim J.; Houle S.; Strafella A. P. Microglial activation in Parkinson’s disease using [18 F]-FEPPA. J. Neuroinflammation 2017, 14, 8. 10.1186/s12974-016-0778-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Holtzman D. M. Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat. Rev. Immunol. 2018, 18 (12), 759–772. 10.1038/s41577-018-0051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley M. E.; Baloh R. H. Inflammation in ALS/FTD pathogenesis. Acta Neuropathol. 2019, 137 (5), 715–730. 10.1007/s00401-018-1933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.; Lüth H. J.; Deuther-Conrad W.; Dukic-Stefanovic S.; Gasic-Milenkovic J.; Arendt T.; Münch G. Advanced glycation endproducts co-localize with inducible nitric oxide synthase in Alzheimer’s disease. Brain Res. 2001, 920 (1–2), 32–40. 10.1016/S0006-8993(01)02872-4. [DOI] [PubMed] [Google Scholar]
- Holmquist L.; Stuchbury G.; Berbaum K.; Muscat S.; Young S.; Hager K.; Engel J.; Münch G. Lipoic acid as a novel treatment for Alzheimer’s disease and related dementias. Pharmacol. Ther. 2007, 113 (1), 154–164. 10.1016/j.pharmthera.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Latta C. H.; Brothers H. M.; Wilcock D. M. Neuroinflammation in Alzheimer’s disease; a source of heterogeneity and target for personalized therapy. Neuroscience 2015, 302, 103–111. 10.1016/j.neuroscience.2014.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q.; Borenstein A. R.; Wu Y.; Jackson J. C.; Larson E. B. Fruit and vegetable juices and Alzheimer’s disease: the Kame project. Am. J. Med. 2006, 119 (9), 751–759. 10.1016/j.amjmed.2006.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beking K.; Vieira A. Flavonoid intake and disability-adjusted life years due to Alzheimer’s and related dementias: a population-based study involving twenty-three developed countries. Public Health Nutr. 2010, 13 (9), 1403–1409. 10.1017/S1368980009992990. [DOI] [PubMed] [Google Scholar]
- Shahidi F.; Ambigaipalan P. Phenolics and polyphenolics in foods, beverages and spices: Antioxidant activity and health effects—a review. J. Funct. Foods 2015, 18, 820–897. 10.1016/j.jff.2015.06.018. [DOI] [Google Scholar]
- Li X.; Han L.; Song Z.; Xu R.; Wang L. Comparative study on the interaction between transferrin and flavonols: experimental and computational modeling approaches. Spectrochim. Acta, Part A 2023, 288, 122128. 10.1016/j.saa.2022.122128. [DOI] [PubMed] [Google Scholar]
- McKay D. L.; Blumberg J. B. A review of the bioactivity and potential health benefits of chamomile tea (Matricaria recutita L.). Phytother. Res. 2006, 20 (7), 519–530. 10.1002/ptr.1900. [DOI] [PubMed] [Google Scholar]
- Gazola A. C.; Costa G. M.; Castellanos L.; Ramos F. A.; Reginatto F. H.; Lima T.; Schenkel E. P. Involvement of GABAergic pathway in the sedative activity of apigenin, the main flavonoid from Passiflora quadrangularis pericarp. Rev. Bras. Farmacogn. 2015, 25, 158–163. 10.1016/j.bjp.2015.03.009. [DOI] [Google Scholar]
- Han J.-Y.; Ahn S.-Y.; Kim C.-S.; Yoo S.-K.; Kim S.-K.; Kim H.-C.; Hong J. T.; Oh K.-W. Protection of apigenin against kainate-induced excitotoxicity by anti-oxidative effects. Biol. Pharm. Bull. 2012, 35 (9), 1440–1446. 10.1248/bpb.b110686. [DOI] [PubMed] [Google Scholar]
- Lee J.-A.; Ha S. K.; Cho E.; Choi I. Resveratrol as a bioenhancer to improve anti-inflammatory activities of apigenin. Nutrients 2015, 7 (11), 9650–9661. 10.3390/nu7115485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza C. S.; Paulsen B. S.; Devalle S.; Lima Costa S.; Borges H. L.; Rehen S. K. Commitment of human pluripotent stem cells to a neural lineage is induced by the pro-estrogenic flavonoid apigenin. Adv. Regener. Biol. 2015, 2 (1), 29244. 10.3402/arb.v2.29244. [DOI] [Google Scholar]
- Rezai-Zadeh K.; Arendash G. W.; Hou H.; Fernandez F.; Jensen M.; Runfeldt M.; Shytle R. D.; Tan J. Green tea epigallocatechin-3-gallate (EGCG) reduces β-amyloid mediated cognitive impairment and modulates tau pathology in Alzheimer transgenic mice. Brain Res. 2008, 1214, 177–187. 10.1016/j.brainres.2008.02.107. [DOI] [PubMed] [Google Scholar]
- Zhao M.; Ma J.; Zhu H.-Y.; Zhang X.-H.; Du Z.-Y.; Xu Y.-J.; Yu X.-D. Apigenin inhibits proliferation and induces apoptosis in human multiple myeloma cells through targeting the trinity of CK2, Cdc37 and Hsp90. Mol. Cancer 2011, 10 (1), 104. 10.1186/1476-4598-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.; Zhang T.; Yang H.; Lan X.; Ying J.; Du G. The flavonoid apigenin protects brain neurovascular coupling against amyloid-β 25–35-induced toxicity in mice. J. Alzheimer’s Dis. 2011, 24 (1), 85–100. 10.3233/jad-2010-101593. [DOI] [PubMed] [Google Scholar]
- Xue B.; DasGupta D.; Alam M.; Khan M. S.; Wang S.; Shamsi A.; Islam A.; Hassan M. I. Investigating binding mechanism of thymoquinone to human transferrin, targeting Alzheimer’s disease therapy. J. Cell. Biochem. 2022, 123 (8), 1381–1393. 10.1002/jcb.30299. [DOI] [PubMed] [Google Scholar]
- Khan S.; Cho W. C.; Hussain A.; Azimi S.; Babadaei M. M. N.; Bloukh S. H.; Edis Z.; Saeed M.; Ten Hagen T. L.; Ahmadi H.; et al. The interaction mechanism of plasma iron transport protein transferrin with nanoparticles. Int. J. Biol. Macromol. 2023, 240, 124441. 10.1016/j.ijbiomac.2023.124441. [DOI] [PubMed] [Google Scholar]
- Hadzhieva M.; Kirches E.; Mawrin C. Review: iron metabolism and the role of iron in neurodegenerative disorders. Neuropathol. Appl. Neurobiol. 2014, 40 (3), 240–257. 10.1111/nan.12096. [DOI] [PubMed] [Google Scholar]
- Shamsi A.; Al Shahwan M.; Ahamad S.; Hassan M. I.; Ahmad F.; Islam A. Spectroscopic, calorimetric and molecular docking insight into the interaction of Alzheimer’s drug donepezil with human transferrin: implications of Alzheimer’s drug. J. Biomol. Struct. Dyn. 2020, 38 (4), 1094–1102. 10.1080/07391102.2019.1595728. [DOI] [PubMed] [Google Scholar]
- Shamsi A.; Ahmed A.; Khan M. S.; Husain F. M.; Amani S.; Bano B. Investigating the interaction of anticancer drug temsirolimus with human transferrin: molecular docking and spectroscopic approach. J. Mol. Recognit. 2018, 31 (10), e2728 10.1002/jmr.2728. [DOI] [PubMed] [Google Scholar]
- Du H.; Xiang J.; Zhang Y.; Tang Y.; Xu G. Structural effects on the conformational transition of transferrin induced by binding of flavonoids with different numbers and positions of hydroxyl groups. J. Photochem. Photobiol., A 2008, 195, 127–134. 10.1016/j.jphotochem.2007.09.014. [DOI] [Google Scholar]
- Shamsi A.; Furkan M.; Khan R. H.; Khan M. S.; Shahwan M.; Yadav D. K. Comprehensive insight into the molecular interaction of rutin with human transferrin: implication of natural compounds in neurodegenerative diseases. Int. J. Biol. Macromol. 2023, 253, 126643. 10.1016/j.ijbiomac.2023.126643. [DOI] [PubMed] [Google Scholar]
- Shamsi A.; Ahmed A.; Khan M. S.; Al Shahwan M.; Husain F. M.; Bano B. Understanding the binding between Rosmarinic acid and serum albumin: in vitro and in silico insight. J. Mol. Liq. 2020, 311, 113348. 10.1016/j.molliq.2020.113348. [DOI] [Google Scholar]
- Huey R.; Morris G. M.; Forli S.. Using AutoDock 4 and AutoDock Vina with AutoDockTools: A Tutorial. The Scripps Research Institute Molecular Graphics Laboratory, 2012.
- Mohammad T.; Mathur Y.; Hassan M. I. InstaDock: a single-click graphical user interface for molecular docking-based virtual high-throughput screening. Briefings Bioinf. 2021, 22 (4), bbaa279. 10.1093/bib/bbaa279. [DOI] [PubMed] [Google Scholar]
- Biovia D. S.Discovery Studio Visualizer: San Diego, CA, USA, 2017; Vol. 936.
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005, 26 (16), 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Wu Y.; Tepper H. L.; Voth G. A. Flexible simple point-charge water model with improved liquid-state properties. J. Chem. Phys. 2006, 124 (2), 024503. 10.1063/1.2136877. [DOI] [PubMed] [Google Scholar]
- Turner P.XMGRACE, Version 5.1.19; Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology: Beaverton, OR, 2005; Vol. 2.
- Mohammad T.; Siddiqui S.; Shamsi A.; Alajmi M. F.; Hussain A.; Islam A.; Ahmad F.; Hassan M. I. Virtual screening approach to identify high-affinity inhibitors of serum and glucocorticoid-regulated kinase 1 among bioactive natural products: combined molecular docking and simulation studies. Molecules 2020, 25 (4), 823. 10.3390/molecules25040823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan M. I.; Anjum D.; Mohammad T.; Alam M.; Khan M. S.; Shahwan M.; Shamsi A.; Yadav D. K. Integrated virtual screening and MD simulation study to discover potential inhibitors of Lyn-kinase: targeting cancer therapy. J. Biomol. Struct. Dyn. 2022, 48, 1–11. 10.1080/07391102.2022.2154849. [DOI] [PubMed] [Google Scholar]
- Noinaj N.; Easley N. C.; Oke M.; Mizuno N.; Gumbart J.; Boura E.; Steere A. N.; Zak O.; Aisen P.; Tajkhorshid E.; et al. Structural basis for iron piracy by pathogenic Neisseria. Nature 2012, 483 (7387), 53–58. 10.1038/nature10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A. A.; Mohammad T.; Hasan G. M.; Hassan M. Advancements in docking and molecular dynamics simulations towards ligand-receptor interactions and structure-function relationships. Curr. Top. Med. Chem. 2018, 18 (20), 1755–1768. 10.2174/1568026618666181025114157. [DOI] [PubMed] [Google Scholar]
- Shukla R.; Tripathi T.. Molecular dynamics simulation in drug discovery: opportunities and challenges. Innovations and Implementations of Computer Aided Drug Discovery Strategies in Rational Drug Design; Springer, 2021; pp 295–316. [Google Scholar]
- Lobanov M. Y.; Bogatyreva N.; Galzitskaya O. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008, 42, 623–628. 10.1134/S0026893308040195. [DOI] [PubMed] [Google Scholar]
- Mohammad T.; Shamsi A.; Anwar S.; Umair M.; Hussain A.; Rehman M. T.; AlAjmi M. F.; Islam A.; Hassan M. I. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: towards the development of effective COVID-19 therapy. Virus Res. 2020, 288, 198102. 10.1016/j.virusres.2020.198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham E.; Dorr B.; Woetzel N.; Staritzbichler R.; Meiler J. Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J. Mol. Model. 2009, 15 (9), 1093–1108. 10.1007/s00894-009-0454-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G. D.; Wolfenden R. Hydrogen bonding, hydrophobicity, packing, and protein folding. Annu. Rev. Biophys. Biomol. Struct. 1993, 22 (1), 381–415. 10.1146/annurev.bb.22.060193.002121. [DOI] [PubMed] [Google Scholar]
- Maisuradze G. G.; Liwo A.; Scheraga H. A. Principal component analysis for protein folding dynamics. J. Mol. Biol. 2009, 385 (1), 312–329. 10.1016/j.jmb.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaleo E.; Mereghetti P.; Fantucci P.; Grandori R.; De Gioia L. Free-energy landscape, principal component analysis, and structural clustering to identify representative conformations from molecular dynamics simulations: the myoglobin case. J. Mol. Graphics Modell. 2009, 27 (8), 889–899. 10.1016/j.jmgm.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Shamsi A.; Mohammad T.; Anwar S.; Nasreen K.; Hassan M. I.; Ahmad F.; Islam A. Insight into the binding of PEG-400 with eye protein alpha-crystallin: multi spectroscopic and computational approach: possible therapeutics targeting eye diseases. J. Biomol. Struct. Dyn. 2022, 40 (10), 4496–4506. 10.1080/07391102.2020.1858964. [DOI] [PubMed] [Google Scholar]
- Ahmad B.; Parveen S.; Khan R. H. Effect of albumin conformation on the binding of ciprofloxacin to human serum albumin: a novel approach directly assigning binding site. Biomacromolecules 2006, 7 (4), 1350–1356. 10.1021/bm050996b. [DOI] [PubMed] [Google Scholar]
- Chaturvedi S. K.; Ahmad E.; Khan J. M.; Alam P.; Ishtikhar M.; Khan R. H. Elucidating the interaction of limonene with bovine serum albumin: a multi-technique approach. Mol. BioSyst. 2015, 11 (1), 307–316. 10.1039/C4MB00548A. [DOI] [PubMed] [Google Scholar]
- Shamsi A.; Amani S.; Alam M. T.; Naeem A. Aggregation as a consequence of glycation: insight into the pathogenesis of arthritis. Eur. Biophys. J. 2016, 45, 523–534. 10.1007/s00249-016-1119-0. [DOI] [PubMed] [Google Scholar]
- Sarzehi S.; Chamani J. Investigation on the interaction between tamoxifen and human holo-transferrin: determination of the binding mechanism by fluorescence quenching, resonance light scattering and circular dichroism methods. Int. J. Biol. Macromol. 2010, 47 (4), 558–569. 10.1016/j.ijbiomac.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Anwar S.; Shamsi A.; Kar R. K.; Queen A.; Islam A.; Ahmad F.; Hassan M. I. Structural and biochemical investigation of MARK4 inhibitory potential of cholic acid: towards therapeutic implications in neurodegenerative diseases. Int. J. Biol. Macromol. 2020, 161, 596–604. 10.1016/j.ijbiomac.2020.06.078. [DOI] [PubMed] [Google Scholar]
- Khan M. S.; Shahwan M.; Shamsi A.; Alhumaydhi F. A.; Alsagaby S. A.; Al Abdulmonem W.; Abdullaev B.; Yadav D. K. Elucidating the interactions of fluoxetine with human transferrin employing spectroscopic, calorimetric, and in silico approaches: implications of a potent Alzheimer’s drug. ACS Omega 2022, 7 (10), 9015–9023. 10.1021/acsomega.2c00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The information supporting this study is available in this article. All the generated data is presented in the manuscript.