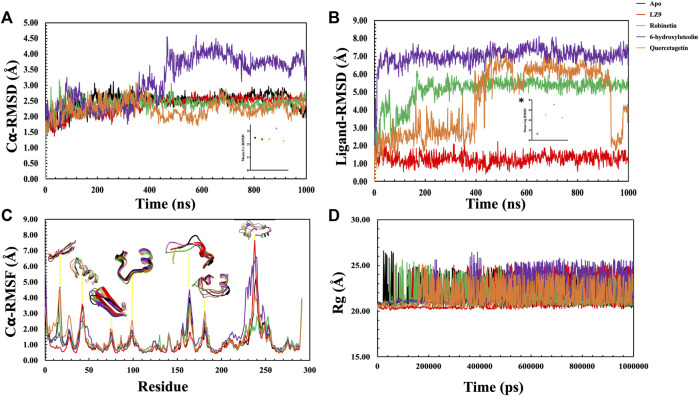
FIGURE 6.
Molecular dynamics simulation trajectories of CDK1 in presence of ligands and absence of ligand (Apo) as a function of 1,000 ns simulation time. (A) Root Mean Square Deviation (RMSD) of Cα atoms backbone. Insert, # shows the mean Cα RMSD values. (B) Root Mean Square Deviation of the ligands with respect to CDK1 protein. The plot displays the RMSD of the ligands after measuring the RMSD of the heavy atoms in the ligand and aligning the protein-ligand complex on the reference protein backbone. If the reported values are noticeably greater than the protein’s RMSD, the ligand has probably diffused away from its initial binding site. Insert, * shows the mean RMSD values. (C): Root Mean Square Fluctuation (RMSF) per residue over time. (D): Radius of gyration for the Apo state and CDK1-ligand complexes.