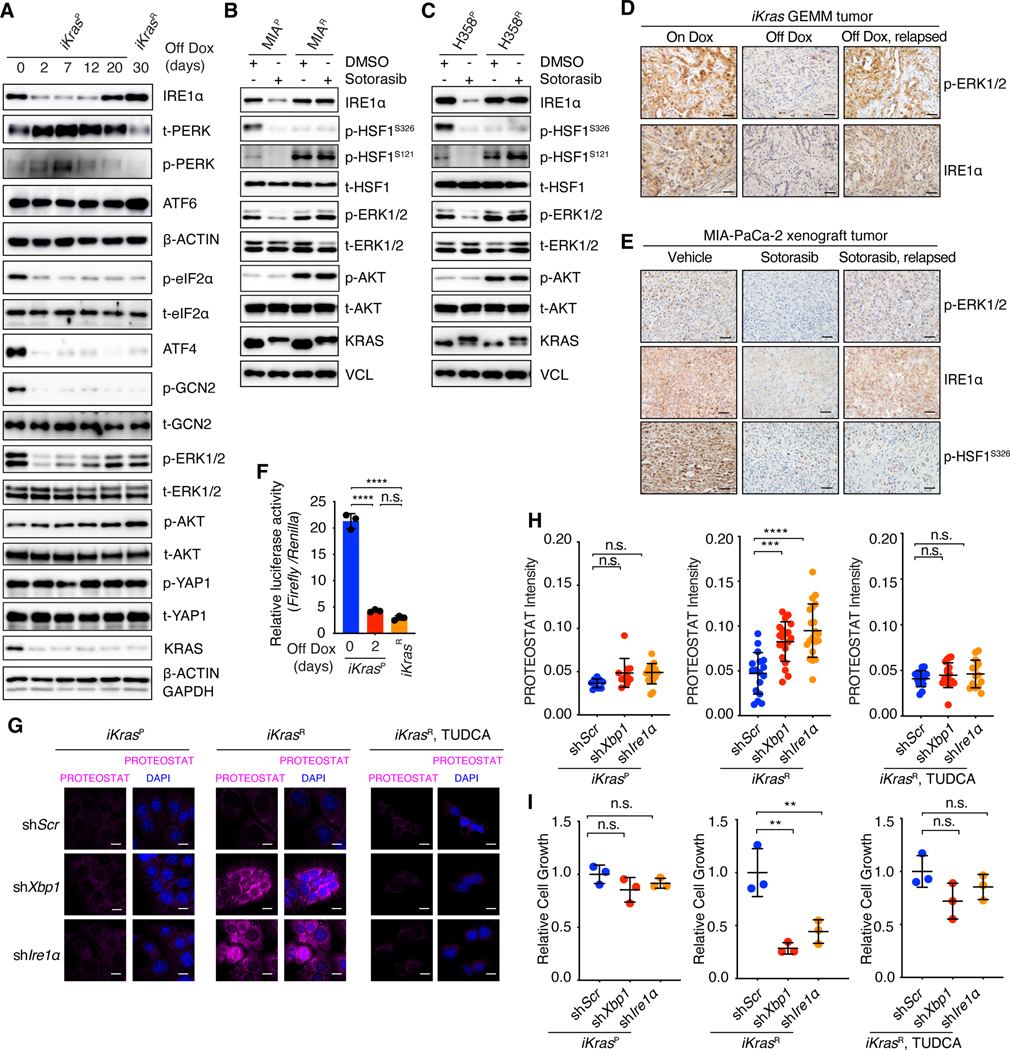
Fig. 2. Oncogenic KRAS inactivation differentially impacts the key nodes of the proteostasis regulatory network.
(A) Immunoblot with indicated antibodies in whole-cell lysates of iKrasP at different time points after KrasG12D inactivation by Dox-withdrawal (Off Dox) until the cells acquired resistance to KrasG12D inactivation (iKrasR cell). (B and C) Immunoblot with indicated antibodies in whole-cell lysates of parental or sotorasib-resistant MIA-PaCa-2 (B) or H358 (C) cells treated with DMSO or 30 nM sotorasib. (D) Immunohistochemical (IHC) staining with indicated antibodies in iKras GEMM tumors treated with doxycycline (On Dox), Dox withdrawal for 3 days (Off Dox), or relapsed after 30 weeks of Dox-withdrawal (Off Dox, relapsed). (E) IHC staining with indicated antibodies in MIA-PaCa-2 xenograft tumors treated with vehicle, sotorasib (30mg/kg for 1 day), or relapsed after 9 weeks of sotorasib treatment (30mg/kg). (F) Relative HSE luciferase activity in iKrasP or iKrasR cells cultured in the presence or absence of Dox for 2 days. Data are shown as mean ± SD, n=3. (G and H) Representative images (G) and quantification (H) of PROTEOSTAT (magenta) and DAPI (blue) staining in iKrasP or iKrasR cells infected with lentiviruses encoding scramble shRNA (shScr), Xbp1 shRNA (shXbp1) or Ire1 α shRNA (shIre1α). Cells were treated with 2.5mM TUDCA dissolved in water for 2 days as indicated. Data represent the average fluorescence intensity of PROTEOSTAT/cell from each image acquired and presented as mean ± SD from n=10 (On Dox), n=17 (Off Dox), or n=17 (Off Dox + TUDCA) images. (I) CCK-8 assay was used to quantify cell viability of iKras cells treated as in G and H. Data are presented as mean ± SD relative to shScr, n=3. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test (H and I) or ordinary one-way ANOVA with Tukey’s multiple comparisons test (F) was used to calculate P values. n.s., not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001. Scale bar: 40μm (D and E) or 20μm (G).