Figure 5.
A specific Lys in the HCDR3 of 1210-like mAbs mediates cross-reactivity to junctional motifs
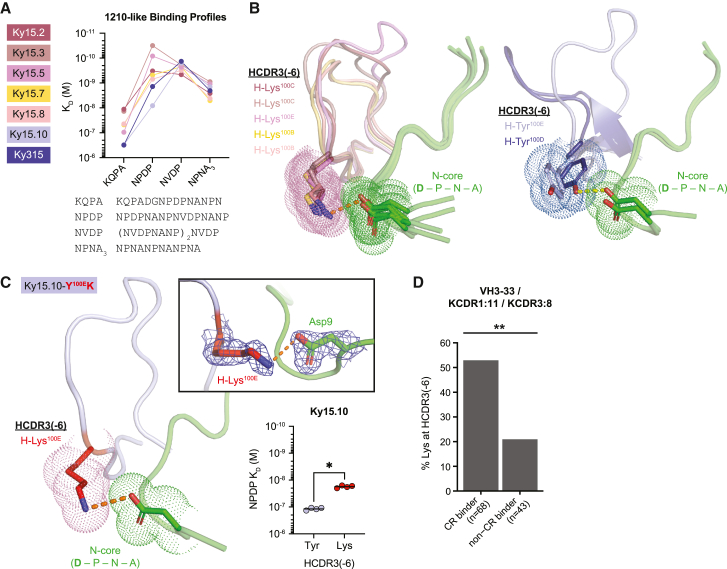
(A) SPR binding profiles of 1210-like mAbs to the indicated peptides derived from the PfCSP junction and repeat regions. Binding profiles are colored by mAb according to the legend on the left, and peptide sequences are listed below. Data represent the mean of three independent measurements.
(B) Non-covalent interactions facilitated by Lys and Tyr residues at position HCDR3(-6). Fab HCDR3s are colored by mAb based on the legend in (A), and peptides are shown in green. Interacting residues are depicted as stick representation with surrounding dots. Salt bridges are indicated by the orange dashed line, and the Ky315-DND HB is shown as a yellow dashed line.
(C) X-ray crystal structure of Ky15.10-Y100EK in complex with the KQPA peptide. Fab and peptide are colored as in (B), with mutated residue shown in red. The top right inset highlights the salt bridge (orange dashed line) between mutated H-Lys100E and peptide Asp9. The composite omit map electron density associated with the salt-bridging residues is shown as blue mesh and contoured at 1.0 sigma. Bottom right: Ky15.10 (HCDR3(-6) Tyr, light blue symbols) and Ky15.10-Y100EK (HCDR3(-6) Lys, red symbols) binding affinity to NPDP peptide as measured by SPR. Symbols represent independent measurements, and the black bar indicates geometric mean. Statistical significance was determined by two-tailed Mann-Whitney test (n = 4): ∗p < 0.05.
(D) Frequency of Lys at position HCDR3(-6) among VH3-33/KCDR1:11/KCDR3:8 mAbs. n indicates the number of mAbs. Cross-reactive (CR) binders were defined as mAbs that bound at least three peptides by ELISA (Figure S1F). Statistical significance was determined using two-proportion Z test: ∗∗p < 0.01.