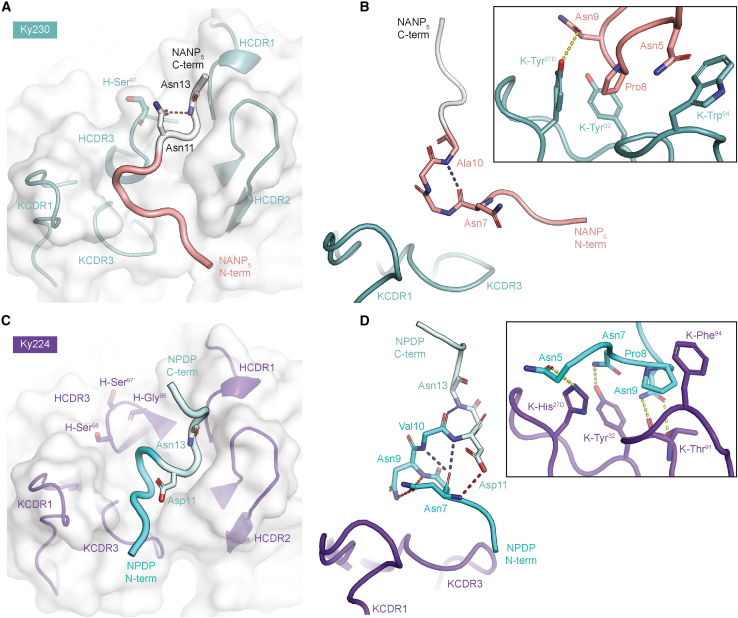
Figure 7.
PfCSP peptides adopt unique conformations when bound to Ky230 and Ky224 mAbs
(A) X-ray crystal structure of Ky230 Fab (white surface, teal cartoon) in complex with NANP5 (N-core colored salmon, C-core colored light gray). CDRs involved in binding, peptide termini, and notable residues are labeled.
(B) Ky230-bound NANP5 N-core conformation with surrounding KCDR1 and KCDR3 loops. The inset highlights interactions contributed by aromatic KCDR1 and KCDR3 residues.
(C) X-ray crystal structure of Ky224 Fab (white surface, purple cartoon) in complex with NPDP (N-core colored cyan, C-core colored pale cyan). CDRs involved in binding, peptide termini, and notable residues are labeled.
(D) Ky224-bound NPDP N-core conformation with surrounding KCDR1 and KCDR3 loops. The inset highlights interchain HBs and CH/π interactions contributed by KCDR1 and KCDR3 residues.
(A–D) Intrachain HBs between main-chain atoms are indicated by blue dashed lines, and those mediated by side-chain atoms are colored red. Fab-peptide HBs are shown as yellow dashed lines.