ABSTRACT
Carbapenems are considered last-resort antibiotics for the treatment of infections caused by multidrug-resistant Enterobacterales, but carbapenem resistance due to acquisition of carbapenemase genes is a growing threat that has been reported worldwide. Klebsiella pneumoniae carbapenemase (blaKPC) is the most common type of carbapenemase in Canada and elsewhere; it can hydrolyze penicillins, cephalosporins, aztreonam, and carbapenems and is frequently found on mobile plasmids in the Tn4401 transposon. This means that alongside clonal expansion, blaKPC can disseminate through plasmid- and transposon-mediated horizontal gene transfer. We applied whole genome sequencing to characterize the molecular epidemiology of 829 blaKPC carbapenemase-producing isolates collected by the Canadian Nosocomial Infection Surveillance Program from 2010 to 2021. Using a combination of short-read and long-read sequencing, we obtained 202 complete and circular blaKPC-encoding plasmids. Using MOB-suite, 10 major plasmid clusters were identified from this data set which represented 87% (175/202) of the Canadian blaKPC-encoding plasmids. We further estimated the genomic location of incomplete blaKPC-encoding contigs and predicted a plasmid cluster for 95% (603/635) of these. We identified different patterns of carbapenemase mobilization across Canada related to different plasmid clusters, including clonal transmission of IncF-type plasmids (108/829, 13%) in K. pneumoniae clonal complex 258 and novel repE(pEh60-7) plasmids (44/829, 5%) in Enterobacter hormaechei ST316, and horizontal transmission of IncL/M (142/829, 17%) and IncN-type plasmids (149/829, 18%) across multiple genera. Our findings highlight the diversity of blaKPC genomic loci and indicate that multiple, distinct plasmid clusters have contributed to blaKPC spread and persistence in Canada.
KEYWORDS: plasmid, carbapenemase, antimicrobial resistance, surveillance studies
INTRODUCTION
Carbapenems are considered last-resort antibiotics for the treatment of infections caused by multidrug-resistant Gram-negative bacteria. Following the use of carbapenems in clinical practice, the emergence of carbapenem-resistant pathogens poses a great threat to human health (1). Carbapenem-resistant Enterobacterales have been reported worldwide as a consequence largely of the acquisition of carbapenemase genes (2).
Of the different classes of carbapenemases, Klebsiella pneumoniae carbapenemase (blaKPC) is the most commonly identified in many countries including the United States and Canada [reviewed in references (3–6)]. blaKPC was the dominant type of carbapenemase isolated from 2010 to 2014 by the Canadian Nosocomial Infection Surveillance Program (CNISP), where they comprised 50%–83% of all carbapenemases analyzed per year (6). From 2017 to 2021, the rates of carbapenemase-producing Enterobacterales infections in Canada increased by 166% (from n = 20 in 2017 to n = 55 in 2021), and blaKPC remains the most common carbapenemase type detected in Canada (7).
Since the first identification of blaKPC almost 30 years ago (8), blaKPC has been found in over 100 different K. pneumoniae sequence types (STs) (9), but the original dissemination was driven primarily by the spread of blaKPC-producing K. pneumoniae isolates that are members of clonal complex 258 (9–12). Along with clonal expansion, blaKPC can disseminate through plasmid- and transposon-mediated horizontal gene transfer and has been detected in at least 11 other genera (3, 10, 13–16).
Advances in genome sequencing have enabled detailed characterization of complete blaKPC-encoding plasmids in recent years (13, 15). blaKPC is associated with a variety of plasmid types, including the narrow-host-range IncF-type plasmids which were crucial for the success of K. pneumoniae clonal complex 258 (3, 12, 15). blaKPC has also been found on broad-host-range plasmids with IncN, IncR, IncX, ColRNA, IncA/C, or IncP replicons around the world (3, 13, 15, 17–20). Many of these plasmids can have multiple replicons, undergo large rearrangements, and encode other genetic features that ensure their persistence (20).
Here, we applied whole genome sequencing to characterize the molecular epidemiology of blaKPC carbapenemase-producing isolates collected by the Canadian Nosocomial Infection Surveillance Program from 2010 to 2021. Using combined short-read and long-read sequencing of selected representatives to generate complete blaKPC-encoding plasmids, we investigated the diversity of carbapenemase-encoding plasmids among these isolates across Canada and compared them to the global context of blaKPC.
RESULTS
Characteristics of blaKPC carbapenemase-producing isolates
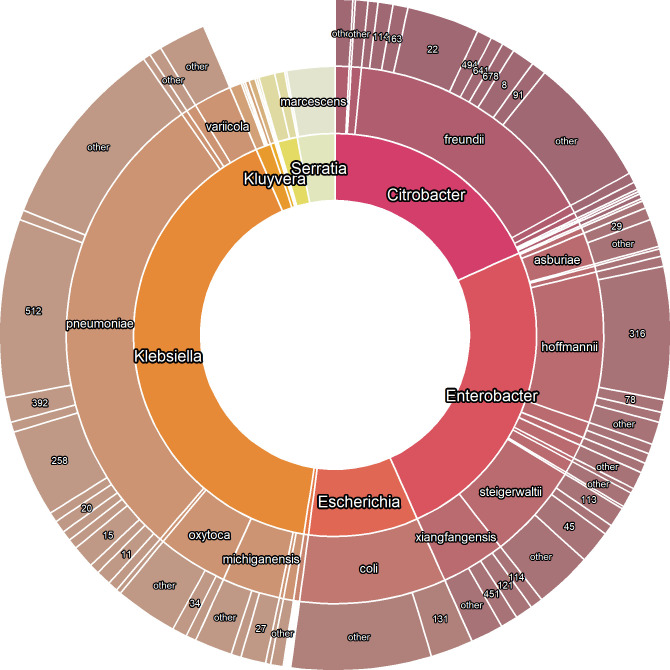
A total of 829 blaKPC-producing isolates were submitted by 34 hospital sites across Canada from 2010 to 2021. We performed short-read and long-read sequencing on a selection of 155 isolates. The blaKPC-producing isolates belonged to 10 genera and 26 different species, with the most common genera being the Klebsiella pneumoniae species complex (268/829, 32.3%), Enterobacter cloacae complex (203/829, 24.4%), Citrobacter freundii complex (132/829, 15.9%), and Escherichia coli (73/829, 8.8%) (Fig. 1). The most common sequence types were K. pneumoniae ST512 (71/268, 26.4%) and ST258 (35/268, 13.1%) (note that ST512 is a single locus variant of ST258), E. cloacae ST316 (53/203, 26.1%), C. freundii ST22 (29/132, 32.0%), and E. coli ST131 (17/73, 23.2%) (Fig. 1).
Fig 1.
Summary of genera (inner ring), species (middle ring), and multi-locus sequence types (MLST; outer ring) of blaKPC-encoding isolates included in the study (829 total isolates). MLST profiles found in three or fewer isolates were grouped into “other”. Not all labels are displayed.
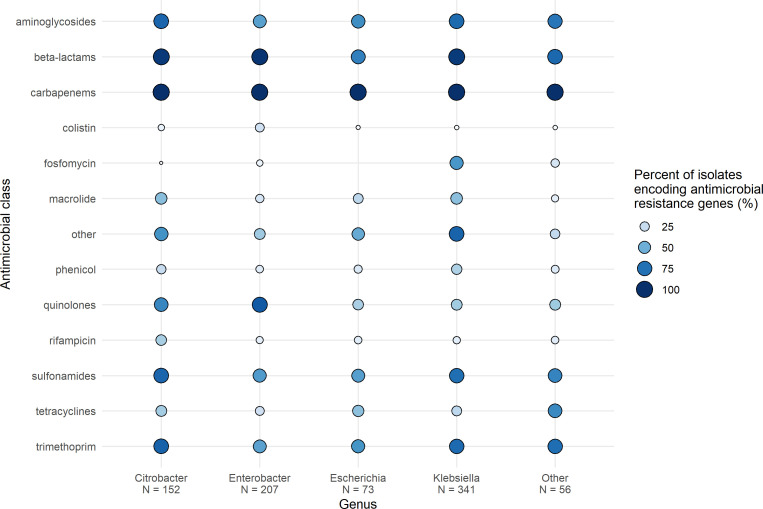
Using the StarAMR tool for the detection of resistance genes in the whole genome sequencing data, we observed 93% (773/829) of blaKPC-producing isolates harbored additional β-lactamase genes alongside the blaKPC carbapenemase (Fig. 2). Of the 773 isolates harboring additional β-lactamases, blaTEM-1B (187/773, 24.2%), blaOXA-9 (146/773, 18.9%), blaTEM-1A (125/773, 16.2%), blaSHV-182 (91/773, 11.8%), and blaOXA-1 (82/773, 10.6%) were the most common types. Aminoglycoside, sulfonamide, and trimethoprim resistance genes were commonly observed among multiple genera (Fig. 2); the most common genes included sul1 (482/829, 58.1%), qacE (424/829, 51.1%), aac(6′)-Ib-cr (357/829, 43.1%), sul2 (227/829, 27.4%), aadA2 (225/829, 27.1%), mphA (224/829, 27.0%), and OqxA/B (224/829, 27.0%). β-Lactamases were significantly more likely to be found in Enterobacter spp., Citrobacter spp., or Klebsiella spp. than E. coli (P < 0.001 for all comparisons). Colistin and quinolone resistance genes were significantly more likely to be found in Enterobacter spp. than any other genera (P < 0.001 for all comparisons). Aminoglycoside, macrolide, sulfonamide, and trimethoprim resistance genes were significantly more likely to be found in Klebsiella spp. or Citrobacter spp. compared to Enterobacter spp. or E. coli (P < 0.035 for all comparisons). Rifampin resistance genes were significantly more likely to be found in Citrobacter spp. than any other genera (P < 0.001 for all comparisons).
Fig 2.
Proportion of isolates encoding antimicrobial resistance genes identified by StarAMR, then categorized by drug class is presented across the top four genera. Values represent proportion of isolates encoding genes belonging to a certain antimicrobial class. “N” indicates the number of isolates in that genus. The “Other” genera include Serratia spp., Raoultella spp., Pseudescherichia spp., Pantoea spp., Morganella spp., Kluyvera spp., and Hafnia spp.
A total of 844 blaKPC genes were detected among the 829 isolates, indicating an occurrence of 1.7% of isolates harboring >1 copy of blaKPC. The blaKPC-3 variant was the most common (678/844, 80.3%) followed by blaKPC-2 (162/844, 19.2%). Two isolates had blaKPC-4, one isolate had blaKPC-9, and one isolate had blaKPC-18. In addition to blaKPC, several genomes carried additional carbapenemase genes (7/829, 0.8%; 2 blaNDM-1, 2 blaNDM-5, 1 blaOXA-232, and 2 blaVIM-1).
Genomic context of blaKPC genes
The blaKPC genes are often located on the Tn4401 transposon which facilitates their mobility to other DNA elements and strains (21, 22). In this data set, blaKPC was frequently found within the previously described Tn4401b isoform (643/829, 77.6%), followed by the Tn4401a isoform (132/829, 15.9%), Tn4401d (5/829, 0.6%), and Tn4401e (1/829, 0.01%). The remainder of blaKPC (38/829, 4.6%) were found on a combination of truncated Tn4401, a unique isoform of Tn4401 or complete absence of Tn4401.
There were 25 different Tn4401 isoforms in this data set. The majority of the single nucleotide variants (SNVs) within Tn4401 observed here differed from those included in TETyper (23), so we generated a custom numbering scheme described in Table S2. The most common SNV profiles were 3 (439/829, 53.0%; A7029G|C8015T), 1 (128/829, 15.4%; none), 2 (115/829, 13.9%; C8015T, differentiates blaKPC-2 and blaKPC-3), 7 (56/829, 6.8%; C8015T|G9882T), 8 (18/829, 2.2%; A7029G|C7672T|C8015T), and 4 (10/829, 1.2%; A7029G|C8015T|T9317C) (Fig. 3).
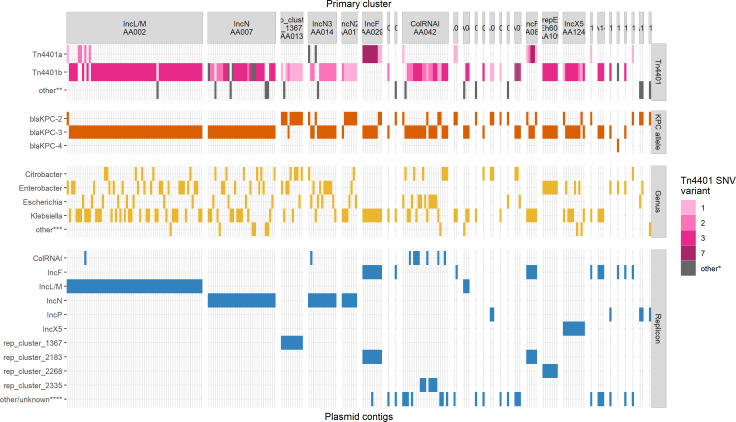
Fig 3.
Characteristics of 202 blaKPC-encoding plasmids sequenced in this study. Groups on the x-axis correspond to primary cluster IDs generated among the plasmid database PLSDB and Canadian blaKPC-encoding plasmids. “Other*” indicates SNVs found in fewer than three plasmids or Tn4401 absence. “Other**” indicates either truncated, alternative isoforms or absence of Tn4401. “Other***” indicates Hafnia, Kluyvera, Morganella, Raoultella, or Serratia genera. “Other/unknown****” indicates replicons (n = 26) found in fewer than five plasmids or replicons that could not be typed.
Complete and circular blaKPC plasmids in a global context
Unsurprisingly, we obtained only 59 closed circular plasmids from Illumina-only assemblies. Using a hybrid approach on a subset of 155 isolates, we obtained an additional 143 complete circular plasmids. Complete blaKPC-encoding plasmids (n = 202) ranged from 7.6 kb to 374.7 kb in size and were distributed among the top species as described above (Fig. 3). Plasmid incompatibility groups included IncL/M (65/202, 32.2%), IncN types (51/202, 25.2%), IncF types (39/202, 19.3%), IncX5 (10/202, 5.0%), and ColRNAI types (21/202, 10.3%), among others. Plasmids were predicted to be conjugative (148/202, 73.2%), mobilizable (36/202, 17.8%), or non-mobilizable (18/202, 8.9%) based on the presence of self-encoded oriT sequences, relaxases, and mating pair formation (MPF) proteins detected by MOB-suite (24).
To investigate where these plasmids fit within a global data set, we clustered our 202 complete plasmids alongside 34,513 plasmids present in PLSDB (25) using the MOB-cluster tool from MOB-suite (24, 26). Altogether, the plasmids clustered into 10,996 distinct primary clusters, and our blaKPC plasmids grouped into a subset of 28 primary clusters which all contained representative plasmids from PLSDB.
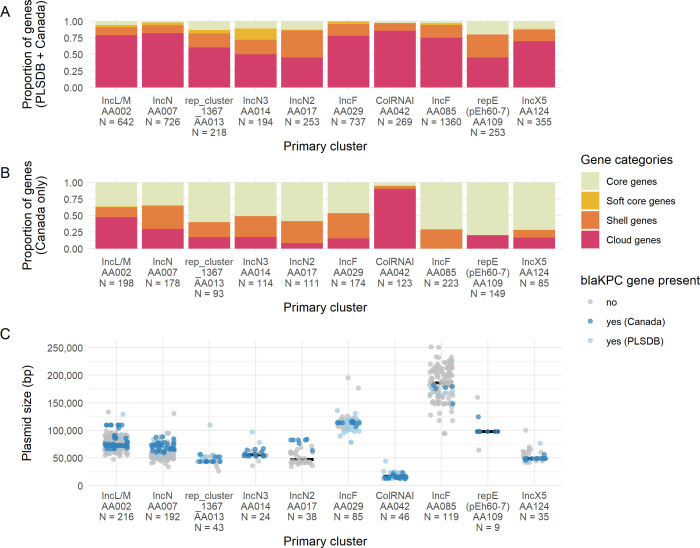
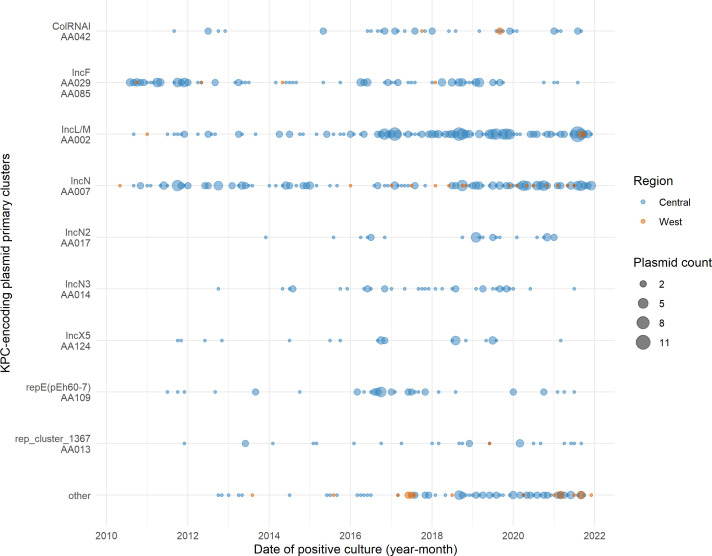
From the above data, we observed 10 primary clusters which represented 87% (175/202) of the Canadian blaKPC plasmids from this study. The primary clusters generally reflected incompatibility groups and the major trends of each are summarized in Table 1. Within the global plasmid data set, the presence of blaKPC varied between our top primary clusters (Fig. 4); some primary clusters tended to be frequently associated with blaKPC (AA014, AA013, AA029, and AA109), whereas others appeared to encode blaKPC more sporadically (AA085 and AA017). Some clusters had a tight distribution of plasmid sizes (AA013 and AA042) whereas others had a broader size distribution (AA085 and AA002). The percentage of core/soft core genes (defined as genes found in >95% plasmids) varied from 3% (AA042) to 28% (AA014) across the primary clusters, indicating some tend to have more diverse or dynamic gene content than others. We also evaluated the core genome size of the Canadian plasmids within each of these primary clusters as we suspected that the Canadian plasmids would be more closely related to each other than to global plasmids. As expected, the percentage of core genes within the Canadian plasmids was higher than the percentage predicted in the global data set with one exception (AA042) (Fig. 4). Several primary clusters had an average of five or more antimicrobial resistance genes (AA002, AA017, and AA085). Certain primary clusters had broad host ranges (AA002, AA007, and AA124), whereas others were only found in a single genus (AA085 and AA109). The dominant type of transposon was Tn4401b except for the IncF clusters (AA085 and AA029) where Tn4401a was dominant.
TABLE 1.
Summary features of top 10 Canadian blaKPC-encoding plasmid clusters among PLSDB
| Primary cluster ID | No. Canadian plasmids | Median size (kb) | Predicted mobilitya,b | Replicon typea,b | Relaxase typea,b | MPF typea,b | No. genera | Core genes (%)d | blaKPC allelea | blaKPC frequency | Tn4401 varianta | Mean no. ARGs per plasmidc | Plasmid taxonomic unit (PTU)e |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA002 | 62 | 75 | Conjugative | IncL/M | MOBP | MPF_I | 12 | 0.09 | blaKPC-3 | 0.34 | Tn4401b | 5.1 | L/M |
| AA007 | 31 | 57.3 | Conjugative | IncN | MOBF | MPF_T | 15 | 0.06 | blaKPC-3 | 0.38 | Tn4401b | 6.1 | N1 |
| AA013 | 10 | 43.6 | Mobilizable | Rep_cluster_1367 | MOBC | −f | 8 | 0.18 | blaKPC-2 | 0.86 | Tn4401b | 2.5 | E8 |
| AA014 | 13 | 55.6 | Conjugative | IncN | MOBF | MPF_T | 4 | 0.28 | blaKPC-3 | 0.71 | Tn4401b | 1.4 | N2/3 |
| AA017 | 7 | 47 | Conjugative | IncN | MOBF | MPF_T | 5 | 0.14 | blaKPC-2 | 0.18 | Tn4401b | 5 | N2/3 |
| AA029 | 9 | 113.6 | Conjugative | IncFIB, IncFII, rep_cluster_2183 | MOBF, MOBF | MPF_F | 4 | 0.04 | blaKPC-2 | 0.86 | Tn4401a | 3.6 | FK |
| AA042 | 21 | 16.5 | Mobilizable | ColRNAI_rep_cluster_1857 | MOBC | − | 7 | 0.03 | blaKPC-3 | 1 | Tn4401b | 1.7 | Could not be assigned |
| AA085 | 185.7 | Conjugative | IncFIB, IncFII, rep_cluster_2183 | MOBF, MOBF | MPF_F | 1 | 0.06 | blaKPC-2 | 0.12 | Tn4401a | 5.9 | FK | |
| AA109 | 7 | 97.9 | Non-mobilizable | Rep_cluster_2268 [novel repE(pEh60-7)] | − | MPF_F | 1 | 0.2 | blaKPC-3 | 0.78 | Tn4401b | 1 | Could not be assigned |
| AA124 | 10 | 48.8 | Conjugative | IncX5 | MOBP | MPF_T | 9 | 0.13 | blaKPC-3 | 0.4 | Tn4401b | 1.6 | New (putative) PTU |
Values indicate the most common genotype in the cluster and may not apply to all plasmids in the cluster.
Values obtained from MOB-suite (24, 26). Mobility is assigned based on the presence of relaxase (mobilizable) and/or MPF proteins (conjugative) or absence of both (non-mobilizable).
ARGs = antimicrobial resistance genes.
Core genes represent the number of genes present in >95% of plasmids in the cluster, divided by the total number of non-redundant genes in the cluster.
PTU values obtained from COPLA (27) .
"-" indicates no relaxase or MPF protein detected.
Fig 4.
Pangenome size and blaKPC prevalence among top 10 MOB-suite primary clusters identified among PLSDB and Canadian blaKPC-encoding plasmids. Pangenome size was calculated for (A) Canadian and PLSDB plasmids, and (B) Canadian plasmids only. Gene categories represent genes found in 99%–100% of plasmids (core), 95%–99% of plasmids (soft core), 15%–95% of plasmids (shell), and 0%–15% of plasmids (cloud). (C) Plasmid length in base pairs and prevalence of blaKPC among plasmids in each primary cluster.
Not surprisingly, all primary clusters with the exception of AA042 contain features that support their stability and persistence in the host cell. Of the genes found in >95% of plasmids in each cluster, between one and five genes (representing 2%–33% of genes per cluster) are involved in stability/transfer/defense which includes genes such as partition/stability genes (e.g., parA, parM, and stbB), anti-restriction genes (e.g., ardA, ardB, and ardR), and SOS-inhibition genes (e.g., psiA and psiB). Different primary clusters had different proportions of stability/transfer/defense gene content; some harbored a single anti-restriction protein (AA124), whereas some encoded two anti-restriction proteins, a plasmid stability protein, an endonuclease, and a stability protein (AA029).
Canadian blaKPC-encoding plasmids in the top primary clusters
Given the high proportion of conserved genes among Canadian plasmids within each primary cluster (Fig. 4), we focused on the features of these Canadian plasmids separately from the other PLSDB plasmids present in each primary cluster. The primary clusters containing the most blaKPC-encoding Canadian plasmids were IncL/M replicons (AA002) and IncN replicons (AA007, AA014, and AA017) (Table 1) and are examined in more detail below.
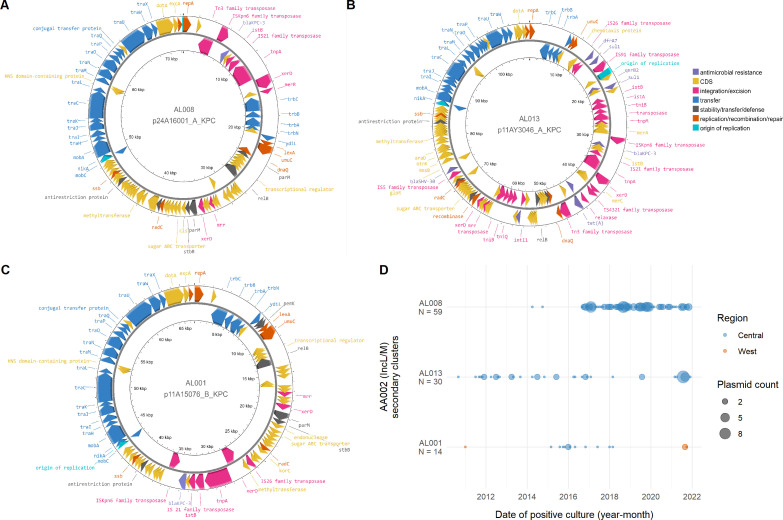
The Canadian plasmids in primary cluster AA002 (IncL/M) were classified into three secondary clusters: AL008 (49/62, 79%), AL013 (8/62, 13%), and AL001 (1/62, 1.6%) (Fig. 5). The first secondary cluster (AL008) had >99% nucleotide identity. They were 72.3 kb isolated from K. pneumoniae species complex, K. aerogenes, K. oxytoca, C. freundii complex, E. cloacae complex, E. coli, and Raoultella planticola between 2016 and 2020 from three sites in the same province. These plasmids harbor blaKPC-3 on Tn4401b-3 and do not encode any other resistance genes. The next secondary cluster (AL013) was also shared among multiple genera with >99% nucleotide identity. They were 109.8 kb isolated from E. coli, K. pneumoniae species complex, and E. cloacae complex from one site between 2011 and 2021. In addition to blaKPC-3 on Tn4401b-3, this plasmid also encodes ant(2″)-Ia, blaSHV-30, dfrA7, qacE, qnrB2, tetA, and two copies of sul1. Structurally, the secondary clusters have similar backbones of transfer and replication genes (Fig. 5). AL008 and AL013 have Tn4401b-3 inserted on the same strand, but AL013 contains many additional integration/excision mobile elements, antimicrobial resistance genes, and hypothetical coding sequences. The AL001 plasmid is structurally similar to AL008 plasmids, but has a different locus of insertion of Tn4401b-3 on the opposite strand and has a pemKI toxin-antitoxin system. The prevalence of these plasmids among different genera and the presence of conjugation genes (MOBP relaxase and MPF_I mating pair formation protein) suggests that regional horizontal transmission has contributed to these secondary clusters’ persistence.
Fig 5.
Diversity of secondary clusters within primary cluster AA002 (IncL/M). Plasmid structures of (A) secondary cluster AL008, (B) secondary cluster AL013, and (C) secondary cluster AL001 which grouped within the AA002 (IncL/M) primary cluster. Coding sequences are colored by function, and not all coding sequences are labeled. (D) Date of positive culture and region of plasmids from primary cluster AA002 (IncL/M). Date of positive culture was grouped into year-month bins. The Central region represents the Canadian provinces of Ontario and Québec, and the West region represents British Columbia, Alberta, Saskatchewan, and Manitoba. “N” indicates the number of isolates that contained assembled or predicted plasmids in the respective secondary clusters.
Three primary clusters (AA007, AA014, and AA017) were identified as having IncN-type replicons with similar features (Table 1). Each cluster corresponds to a different type of replicon: IncN (AA007), IncN2 (AA017), and IncN3 (AA014). All plasmids in primary cluster AA007 were grouped in a single secondary cluster (31/31, 100%), as did all plasmids in primary cluster AA017 (7/7, 100%); in contrast, plasmids in primary cluster AA014 were split into two secondary clusters: AL059 (11/13, 84%) and AL060 (2/13, 15%). The first secondary cluster (AL059) was isolated from E. coli, K. pneumoniae, and E. cloacae complex from five sites in one province between 2014 and 2020. These plasmids encode blaKPC-3 on Tn4401b-2 and no other resistance genes with the exception of a single plasmid with a multi-resistance gene island. The other secondary cluster (AL060) was found in C. freundii complex and K. pneumoniae complex at two sites in one province. This group has no resistance genes aside from blaKPC-2 on Tn4401a-2. Primary cluster AA017 plasmids were isolated from E. coli, K. pneumoniae complex, E. cloacae complex, and Citrobacter farmeri from two sites in one province between 2016 and 2020. Aside from blaKPC-2 on Tn4401b-1, other resistance genes found in this cluster include blaTEM-1B, dfrA25, mph(A), qacE, qnrB2, and sul1. Plasmids in the largest IncN primary cluster (AA007) were isolated from E. coli, K. pneumoniae complex, Klebsiella oxytoca, Klebsiella michiganensis, Raoultella ornithinolytica, E. cloacae complex, C. freundii complex, Citrobacter amalonaticus, Citrobacter koseri, and Serratia marcescens. These plasmids were isolated from 2012 to 2020 from nine sites in four provinces. This group contains blaKPC-3 on Tn4401b with many of them encoding different combinations of aac(6′)-Ib, aph(3″)-Ib, aadA1, blaOXA-9, blaTEM-1A, dfrA14, qacE, qnrS1, sul1, sul2, and tetD. The prevalence of IncN plasmids in multiple genera from multiple sites in the same province (primary clusters AA017 and AA014) or across multiple provinces (AA007) along with encoded conjugation genes (MOBF relaxase and MPF_T mating pair formation protein) suggests there has been widespread horizontal transmission of these primary clusters in Canada.
Canadian blaKPC-encoding plasmids in the other primary clusters
Plasmids in primary cluster AA042 are small in size (10–20 kb) and grouped into four secondary clusters: AL143 (2/21, 10%), AL144 (17/21, 81%), AL149 (1/21, 5%), and AL150 (1/21, 5%). The largest secondary cluster (AL144) had multiple replicon types identified by MOB-suite, with the most common being ColRNAI_rep_cluster_1987 and rep_cluster_2335. These plasmids were isolated from 14 sites across three provinces from C. freundii complex, E. coli, K. pneumoniae complex, and S. marcescens between 2011 and 2021. These plasmids did not contain other resistance genes and most (14/17, 82%) were classified as “mobilizable” by MOB-suite, which indicates they encoded a relaxase or recognizable oriT sequence. The core genes in this group include all components of the Tn4401b transposon including blaKPC-3 which results in integration/excision genes being common in this secondary cluster. The small size of mobilizable plasmids in this primary cluster and their prevalence at various sites indicate that blaKPC spread may be influenced by these smaller vectors.
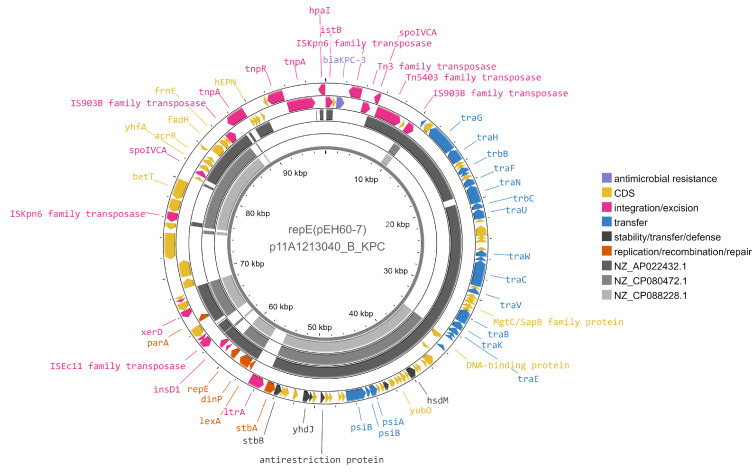
We detected a novel replicon type in primary cluster AA109, designated as rep_cluster_2268 by MOB-typer, which now has a novel replicon designation of repE(pEh60-7) in PlasmidFinder (personal communication). We obtained complete plasmid sequences for seven isolates which grouped into a single secondary cluster. These plasmids were only found in the Enterobacter cloacae species complex, specifically Enterobacter hormaechei subsp. hoffmannii ST316, at a single site in one province from 2011 to 2020, indicating a very stable plasmid or reservoir of this strain. Occurrences of this plasmid are intermittent from 2011 to 2016 and appear to become more prevalent from 2016 to 2018 with fewer detected in recent years. SNVPhyl (28) was run to analyze SNV differences in the ST316 blaKPC-harboring isolates from this site over the 8-year time span (n = 51), and a maximum of 62 SNVs was identified between isolates using 88.51% of the genome. The novel repE(pEh60-7) plasmids encode blaKPC-3 in Tn4401b-3 but did not encode any other resistance genes. While they were classified as non-mobilizable by MOB-suite, they do encode a mating pair formation protein and other genes annotated as conjugation machinery which may not be represented in current databases (Fig. 6). Further work will need to be done to determine if these plasmids are conjugative. These plasmids also encode for stability/transfer/defense proteins including a SOS-inhibition protein, stability partition proteins, the hok-sok toxin-antitoxin system, and an anti-restriction protein. There were two PLSDB plasmids that grouped in the same primary AA109 cluster but in different secondary clusters, and their gene content is quite diverse compared to the Canadian plasmids (70% identity to NZ_AP022432.1 and 43% identity to NZ_CP080472.1) (Fig. 6). Given these plasmids are only found in one specific sequence type, clonal transmission is likely driving this primary cluster’s persistence.
Fig 6.
Comparison of p11A1314040_B_KPC, representative of plasmids with novel replicon repE(pEh60-7) and reference NZ_CP088228.1 (pEh60-7), NZ_CP080472.1 (PLSDB), and NZ_AP022432.1 (PLSDB) from cluster AA109. Reference sequences were aligned using blastn+ and an e-value cut-off of 0.1. NZ_CP080472.1 and NZ_AP022432.1 were placed in the same primary cluster by MOB-cluster as the reference p11A1314040_B_KPC but in different secondary clusters. Genes are colored by functional classes determined by mobileOG-db, and genes without annotations are hypothetical proteins. Not all gene labels are displayed.
Primary cluster AA124 plasmids were classified as IncX5 replicons grouped into two secondary clusters: AL404 (1/10, 10%) and AL405 (9/10, 10%). The largest secondary cluster (AL405) encoded blaKPC-3 on Tn4401b-3. These plasmids were isolated from K. pneumoniae complex, Serratia liquefaciens, E. cloacae complex, and Klebsiella ascorbata from four sites in two provinces between 2015 and 2021. Eight of these nine plasmids are >99% identical and do not contain any resistance genes aside from blaKPC-3 on Tn4401b-3. These plasmids are predicted to be conjugative based on the presence of the MOBP relaxase and MPF_T mating pair formation protein. The wide range of genera suggests that these IncX5 plasmids have disseminated horizontally across the country.
Two primary clusters (AA029 and AA085) contained IncF-type replicons that were found exclusively in K. pneumoniae isolates. All plasmids encoded two MOBF relaxases and an MPF_F mating pair formation protein which suggests that they are conjugative. Plasmids in primary cluster AA085 were grouped into a single secondary cluster and were isolated from two sites in two provinces from 2011 to 2020. These plasmids were from ST512 (3/5, 60%) and ST258 (2/5, 40%) isolates and varied in size (range: 148–182 kb). All plasmids contained Tn4401a, but blaKPC-2 was present in ST258 and blaKPC-3 was present in ST512, and all but one encoded aadA2, aph(3′)-Ia, drfA12, mphA, qacE, and sul1. Primary cluster AA029 plasmids were grouped into three secondary clusters which reflected the sequence type of the host: AL107 (ST512, 6/9, 67%), AL104 (ST258, 2/9, 22%), or AL106 (ST152, 1/9, 11%). Two of these secondary clusters (AL107, ST512; AL106, ST152) encoded blaKPC-3 on Tn4401a-7, whereas the other (AL104, ST258) encoded blaKPC-2 on Tn4401a-1. The secondary clusters AL107 and AL106 (ST512, ST152) were isolated from the same single site from 2010 to 2011, and the ST258 plasmids were isolated from two sites in one province in 2016 and 2019. Both secondary clusters AL107 and AL106 (ST512, ST152) also encoded blaOXA-9 and blaTEM-1A. One of the first described pKpQIL plasmids in K. pneumoniae ST258 [NC_014016.1 (29)] in PLSDB clustered within AA029 (secondary cluster AL107) and also encoded blaOXA-9 and blaTEM-1A. These pKpQIL-type plasmids have been globally disseminated through the expansion of K. pneumoniae clonal complex 258 (9, 29–31), and our results support continued clonal dissemination of these plasmid types in Canada.
Epidemiology of blaKPC-encoding plasmid clusters across Canada
Long-read data are essential for resolving plasmid structures (32), but this can be costly for large surveillance data sets when performed alongside short-read sequencing, which then makes it difficult to analyze plasmid populations at a broad scale. Consequently, we explored MOB-recon (24) as a tool to predict plasmid cluster presence in isolates with incomplete blaKPC-encoding contigs (635/829, 76.6%) using a database containing a subset (~20%) of our isolates that were long-read sequenced. We successfully assigned the majority of blaKPC-encoding contigs to an existing plasmid primary cluster that we defined above, demonstrating that MOB-recon is a feasible approach to predict plasmid cluster membership without long-read sequencing all isolates.
MOB-recon assigned 95% (603/635) of incomplete blaKPC-encoding contigs to an existing primary cluster, despite approximately half of blaKPC contigs (302/635, 48%) not matching any replicon, relaxase, MPF, or oriT sequence in the MOB-suite database. The 10 largest primary clusters contained 89% (539/603) of blaKPC contigs. Similar to the trends observed above (Table 1), the largest number of blaKPC contigs were classified into the IncL/M cluster (AA002, 142/635, 22%) and the IncN clusters (AA007, 149/635, 23%; AA014, 23/635, 3.6%; and AA017, 27/635, 4.3%) (Fig. S1). All contigs assigned to primary cluster repE(pEh60-7) (AA109) (34/635 contigs, 5%) belonged to E. hormaechei subsp. hoffmannii ST316, and all contigs assigned to the IncF primary cluster (representing both AA029 and AA085) (94/635, 15%) were from the K. pneumoniae species complex, which further confirms that these two primary clusters are species-specific. Finally, the remaining contigs were placed in the ColRNAI cluster (AA042, 27/635, 4%) the IncX5 cluster (AA124, 16/635, 3%), and the rep_cluster_1367 (AA109, 27/635, 5%). This reflects the distribution of complete blaKPC plasmid clustering alongside PLSDB as described above (Fig. 4; Table 1). Several other replicon types of blaKPC-encoding contigs were present at low frequencies (21 IncHI2A, 14 IncX3, 12 IncC, and 9 IncP). Those contigs that were unclassified appeared to be chromosomal (located on contigs >1 Mb; 3/32, 9%), coincided with the 10 kb length of Tn4401 and were filtered out (6/32, 19%), or did not match any plasmids currently in the database, indicating that further sequencing is required to confirm the genomic location of the blaKPC in these isolates.
Given the majority of blaKPC-encoding contigs were predicted to be of plasmid origin, we examined the temporal and geographic patterns of all blaKPC-encoding plasmids in Canada from 2010 to 2021 (Fig. 7). Geographically, the incidence of blaKPC-encoding isolates is much higher in the Central region than the West (Central: n = 777/829, 93.7%; West: n = 52/829, 6.3%). The three primary clusters that have persisted over the longest time frame are IncN (AA007), AA002 (IncL/M), and AA0029/AA085 (IncF). Occurrences of AA002 (IncL/M) plasmids were low until 2016, but appear to be found more frequently for the remainder of the study period. Some of the earliest detected plasmids were AA029/AA085 (IncF) plasmids, but these were observed less frequently from 2020 onward, perhaps as AA002 (IncL/M) plasmids increased in prevalence in the Central region. The AA007 (IncN) plasmids appear to be established in the population. Since 2018, there has been an increase in replicons in the “Other” category or those plasmids that do not fall into the top 10 primary clusters, particularly in the West region. This may indicate that other plasmids are becoming dominant in the West region, and further investigation into smaller Western-specific clusters (such as plasmids with IncP6 replicons) is required.
Fig 7.
Epidemiology of Canadian blaKPC-encoding plasmid primary clusters from 2011 to 2021. Date of positive culture was grouped into year-month bins. The Central region represents the Canadian provinces of Ontario and Québec, and the West region represents British Columbia, Alberta, Saskatchewan, and Manitoba. “Other” indicates plasmids which did not group in the top 10 primary clusters.
We examined the epidemiology of the three secondary clusters in primary cluster AA002 (IncL/M) in more detail, as this is the only primary cluster wherein we had a substantial number of plasmids in multiple secondary clusters (Fig. 5D). From an epidemiological perspective, AL013 plasmids have been present for a longer time frame at low abundances with an increase in prevalence in 2021, whereas AL008 plasmids have appeared consistently since 2016. It appears that secondary cluster AL001 may be spreading to the Western region, as this replicon was detected initially in the West in 2010 but not again until 2021 (Fig. 5D). The independent samples collected over long time periods indicate that these plasmids seem to be stably circulating in these regions.
DISCUSSION
We examined the prevalence and distribution of blaKPC-producing Enterobacterales and their plasmids in Canada from 2010 to 2021. Our results suggest that a combination of regional clonal transmission (AA109, AA029, and AA085), regional horizontal transmission (AA017, AA124, AA002, and AA014), widespread horizontal transmission (AA007), and other means of mobilization (AA013 and AA042) has contributed to blaKPC prevalence in Canada.
Here, we report on a novel blaKPC plasmid, repE(pEh60-7), isolated exclusively from E. hormaechei subsp. hoffmannii ST316, which has been stably circulating in this population for over 8 years. There have been limited reports on the E. cloacae complex ST316 lineage elsewhere; one study identified nine E. hormaechei ST316 isolates encoding blaKPC-2 in the United Kingdom between 2014 and 2016 as part of a larger reference study, however, the plasmid content was not reported (33). Otherwise, eight isolates [one each from references (34–36), and five from reference (37)] of E. hormaechei ST316 have been recently been reported to carry blaNDM-1 in Asia. Although conjugation genes were present, the narrow host range of the plasmid suggests that blaKPC persistence is likely due to the ST316 clone and not driven by the plasmid.
We report on some plasmid clusters that are found across a variety of species, supporting evidence of horizontal transfer of blaKPC. One such primary cluster was AA002, which contained the IncL/M plasmids isolated from Citrobacter spp., Escherichia spp., Enterobacter spp., Klebsiella spp., and Raoultella spp. isolates. The secondary cluster AL008 plasmids had a single resistance gene, but secondary cluster AL013 plasmids encoded fluoroquinolone resistance genes, additional β-lactamase genes, and other antimicrobial resistance genes which may favor their persistence in hospital environments. In addition, all IncL/M plasmids encoded a ParB-like partitioning/stability protein, an anti-restriction protein, a restriction endonuclease (Mrr), single-stranded DNA-binding protein (Ssb), and the RelB antitoxin although no RelE toxin was detected in any plasmid sequence. These features likely contribute to their long-term persistence and stability in Canada. However, blaKPC-encoding IncL/M replicons appear to be rare as they are notably absent from other recent blaKPC surveillance studies (13, 15, 38, 39). One study reported that blaKPC-2 being associated with IncL/M replicons a multi-clonal K. pneumoniae outbreak in a region in Argentina, but the relatedness between plasmids was not investigated (40). IncL/M plasmids are typically associated with blaNDM, blaIMP, and blaOXA-type carbapenemases (3, 12, 13, 41–43), although a select few cases have been reported in diverse hosts (19, 44–47). The pNE1280 IncL/M plasmid, which was one of the first IncL/M plasmids described carrying blaKPC-2 (45), clustered in the AL001 secondary cluster, which was not well represented in the Canadian data set. We demonstrated ongoing blaKPC-encoding IncL/M plasmid transfer across multiple genera which merits further investigation of IncL/M-type replicons as potential vectors for long-term blaKPC persistence.
In contrast to the IncL/M plasmids, the IncN plasmid family had the broadest phylogenetic and geographical distribution across Canada, which is unsurprising given their broad host ranges and their long association with blaKPC genes (3, 48, 49). blaKPC-encoding IncN plasmids have been linked to multi-center outbreaks in Columbia (50) and Germany (51), both facilitated by intra- and inter-species horizontal plasmid transfer. Other blaKPC-encoding IncN plasmids implicated in inter-patient transfer grouped in the same secondary clusters as the Canadian plasmids here [plasmid 12 (49) and pYDC107_70 (52) in AA007/AL033; pKPC-SMH (53) in AA014/AL059], indicating high sequence similarity among Canadian plasmids to those found across the globe. Similar to IncL/M plasmids, all IncN plasmids encoded partitioning/stability proteins (StbA, StbC, and StdB), at least two anti-restriction proteins (ArdA, ArdB, and ArdR), an endonuclease (EcoRII and PemK), and other antimicrobial resistance genes. Given these features are also common in IncL/M plasmids, further investigation is required to explain why the IncN plasmids have a broader geographic and host range distribution in Canada than the IncL/M plasmids.
The IncF-type replicons observed here [IncFIB(pQil)/IncFII(K) and IncFIB(K)/IncFII(K)] are common in blaKPC-producing K. pneumoniae species complex isolates detected in other nationwide surveillance programs (12, 13, 15, 17, 18, 38, 39, 54). IncF-type replicons have been implicated in clinical outbreaks of blaKPC-producing K. pneumoniae in the Netherlands (55) and the United States (56) as well as cross-species plasmid transmission in the United Kingdom (57). The clonal lineage ST258/512 accounts for 39.6% (106/268) of blaKPC-producing K. pneumoniae isolates in our collection (Fig. 1) which were isolated across 3 provinces and 12 sites. This frequency is lower than frequencies observed in Colombia [47% (18)], in the United States [74% CRACKLE-2 (5)], and across Europe [73% EuSCAPE (13), 76% Europe/Israel (39)], but is higher than frequencies reported in the United Kingdom [20% (15)] and Argentina [0% (40)]. Our findings support that IncF-type plasmids in K. pneumoniae ST512 and ST258 are important in Canada for blaKPC persistence in the clonal group ST258/512, although there is more diversity in the sequence type of blaKPC-encoding K. pneumoniae isolates in Canada compared to other parts of the world. Interestingly, IncF-type plasmids were less common in recent years (2020 and 2021), indicating other plasmid types such as IncL/M and IncN-type replicons may be replacing them in the Canadian landscape.
Small cryptic plasmids are often overlooked (58) and underrepresented in plasmid-mediated antimicrobial resistance studies (59). ColRNA-type replicons smaller than 22 kb were the third largest blaKPC-encoding primary clusters in our data set. ColRNA-type plasmids were predicted to be found in >90% of carbapenemase-producing Klebsiella pneumoniae isolates across Europe (13, 60) and around the world (61), although these authors did not investigate if any carbapenemase genes were found on these plasmids. Similarly, ColRNAI replicons were present in >50% of blaKPC-producing isolates in Colombia (18). Our MOB-recon results agree with these data and predict that 69% (568/829) of Canadian isolates contain ColRNAI-type or rep_cluster_2335 plasmids, the majority (524/568, 92%) of which do not encode blaKPC (data not shown). These plasmids can be important vectors for transferring antimicrobial resistance genes between isolates, including the transfer of blaKPC-type carbapenemases (19, 31, 62). Experimentally, ColRNAI plasmids encoding blaKPC and Tn4401 could mobilize between strains and participate in Tn4401 transposition events within a patient (63). About half the plasmids we sequenced in this primary cluster do not encode a relaxase but they could be mobilized by the relaxase of a co-resident plasmid if the oriT sequence has enough similarity as shown in a previous study (64). Given their high prevalence and ability to mobilize resistance elements, these small plasmids are likely contributing to blaKPC dissemination and merit further investigation as potential vectors for blaKPC and other antimicrobial resistance spread across the globe.
Plasmid clustering and analyses require a methodology that is tolerant to a small amount of changes given the plasticity of plasmid sequences. Clustering plasmids with MOB-cluster provided a way to analyze subsets of similar plasmids, and MOB-recon allowed us to predict the prevalence of these plasmid clusters in isolates with incomplete assemblies. Canadian plasmids are grouped in two or three secondary clusters within each primary cluster (Mash distance <0.025), with typically a single secondary cluster containing the majority of Canadian plasmids in that primary cluster, indicating many of them have near duplicate sequences (26, 65) and this provided a starting point for investigating clusters of interest in more detail.
Analyzing broad-scale plasmid populations is desirable for surveillance programs, and ideally, every isolate would be sequenced to obtain complete plasmid sequences. Short-read sequencing alone results in many plasmids remaining fragmented (32) but performing long-read sequencing in addition to short-read on large collections of isolates can be cost and time-prohibitive, so we explored using MOB-recon to predict putative plasmids in incomplete assemblies after long-read sequencing a subset of isolates. There is no standardized method for estimating plasmid presence/containment in fragmented assemblies. It is important to recognize that any reference-based method for plasmid reconstruction involving mapping to reference sequences to infer presence assumes that plasmid structures are relatively conserved, and this approach can produce misleading results if plasmid plasticity is high (66, 67). Similar studies have used short-read mappers such as BWA/Bowtie2/SMALT (13, 43, 68, 69) or k-mer-based tools (60, 70) to determine plasmid prevalence in incomplete assemblies, typically using a select few plasmids as references. We use a homology-based clustering approach with MOB-recon (24) to reconstruct plasmids using a relatively large set of complete Canadian plasmids (1,856 plasmids). Previous work has shown that MOB-recon performed well compared to other tools (58, 71, 72), and that Enterobacterales plasmid replicons and mobility genes are well represented in the MOB-suite database (58). Using this approach, we were able to predict plasmids for the majority of incomplete blaKPC-encoding contigs (605/635, 95%). For several primary clusters that were only present in specific sequence types [IncF-type, AA029/AA085 and repE(pEh60-7), AA109], those blaKPC-encoding contigs that were predicted to be part of plasmids in those primary clusters were from isolates with matching sequence types, which further supports this methodology for plasmid analysis in large-scale Enterobacterales surveillance data and eliminates the requirement to perform long-read sequencing on every single isolate.
In summary, there are multiple distinct clusters of blaKPC-encoding plasmids that circulated in Canada from 2010 to 2021. Certain plasmid clusters spread by horizontal transmission and were found in multiple genera in multiple provinces whereas others persisted through clonal dissemination of the host organism. Our findings highlight the need to integrate targeted long-read sequencing into carbapenemase-producing organism surveillance to generate complete plasmid assemblies and demonstrate that plasmid clustering can facilitate analyses of a large number of plasmids. The characterization of blaKPC plasmids is not only important for outbreak control but also for epidemiological surveillance of antimicrobial resistance, as plasmids encoding blaKPC genes typically carry other antimicrobial resistance genes that can be exchanged between species.
MATERIALS AND METHODS
Surveillance period and PCR confirmation of blaKPC carbapenemase gene
CNISP is a sentinel surveillance system which collects epidemiological and linked microbiology data from 90 Canadian acute-care hospitals across 10 provinces and 2 territories. Enterobacterales isolates non-susceptible to a carbapenem (as determined by the submitting site) isolated from patients from 2010 to 2021 were voluntarily submitted to the National Microbiology Laboratory (Winnipeg, Canada) by Canadian hospitals and provincial public health laboratories for carbapenemase gene detection. Multiplex PCR to confirm the carbapenemase gene blaKPC was conducted as previously described (54). A total of 829 isolates encoding blaKPC were collected from 2010 to 2021 from 34 hospitals (Table S1), with one hospital submitting 38% (317/829) of all isolates. Where applicable, the Central region refers to the provinces of Ontario and Québec, and the West region refers to the provinces of British Columbia, Alberta, Saskatchewan, and Manitoba. See the Supplemental Materials for additional information about the surveillance program and isolate eligibility criteria.
Whole genome sequencing
All 829 isolates were sequenced with Illumina MiSeq platforms, and 155 of these were additionally sequenced using Oxford Nanopore Technologies (ONT). Isolates for ONT long-read sequencing represented about 20% of all blaKPC cases stratified across all provinces in Canada. For sites with known outbreaks, we took representative isolates to prevent oversampling of clonal cases. The average Illumina depth of coverage was 123×, and the average ONT depth of coverage was 75×. Genomes were trimmed and filtered before assembly with Unicycler v0.5.0 (73). A total of 111 chromosomes and 1,856 plasmids were completed and circularized in our data set. Details are in the Supplementary Materials, and details on blaKPC-encoding plasmids can be found in Table S2.
Bioinformatic analyses
Organism genus was confirmed using the RefSeq Masher Matches tool (74) and FastANI v1.3 (75) for Enterobacter spp. Species complex definitions are in the Supplementary Materials. StarAMR v0.9.1 (76) was used to detect antimicrobial resistance genes using the ResFinder database v2022-05-24 (77) and sequence type using the multi-locus species types database v2.23.0 (78, 79). TETyper v1.1 (23) was used to identify the Tn4401 isoforms using the reference Tn4401b-1 found in the GitHub repository (accession CP017937.1:29609-39614) and variants using an updated SNP profile file (Table S3) and an updated structural profile file (Table S4) based on the literature (22, 23, 45, 80–84). Panaroo v1.3.2 (85) was used to estimate the pangenome and generate core gene alignments for each primary cluster as well as alignments for each of the top four genera (Citrobacter, Enterobacter, Escherichia, and Klebsiella). The MOB-typer tool from MOB-suite v3.1.4 (24, 26) was used to identify plasmid replicons and mobility classes using the default databases.
Plasmid clustering and containment analysis
MOB-suite primary cluster designations are a useful way to broadly cluster plasmids for epidemiological studies, and so plasmids assigned to different primary MOB-clusters are sufficiently unrelated to not be considered as part of an epidemiologically relevant transmission event (26, 65). However, plasmids that share the same primary cluster designation can be examined in more detail through higher resolution subtyping such as secondary cluster designations. If two plasmids are assigned to the same secondary cluster, they have near duplicate sequences and are sufficiently related to be strong candidates for outbreak investigations (26, 65). In addition to secondary cluster designation, epidemiological data are required to best assess direct plasmid transmission.
For plasmid clustering analysis, the PLSDB v2021_06_23v2 (25) database was downloaded and clustered alongside the 202 circular blaKPC-encoding plasmids in this study using MOB-cluster from the MOB-suite v3.1.4 package (24, 26). For plasmid containment analysis, all 1,856 circular plasmids completed in this study (including the 202 blaKPC-encoding plasmids) were clustered using MOB-cluster to create a custom Canadian plasmid database. All 829 isolates were screened for plasmids with MOB-recon using this custom database, and the output was filtered to focus on the reconstructed plasmids containing blaKPC. Further details on plasmid clustering and containment are provided in the Supplementary Materials.
ACKNOWLEDGMENTS
We thank the physicians, epidemiologists, infection control practitioners, and laboratory staff at each participating hospital for their contributions to the study. We gratefully acknowledge the Genomics Core Facility of the National Microbiology Laboratory, Public Health Agency of Canada for whole genome Illumina sequencing, the Bioinformatics Core Facility of the National Microbiology Laboratory, Public Health Agency of Canada, for computational infrastructure, and the Robotics Core Facility, National Microbiology Laboratory, Public Health Agency of Canada, for DNA extractions for Illumina sequencing.
The Public Health Agency of Canada provided funding for the Canadian Nosocomial Infection Surveillance Program.
Contributor Information
Laura Mataseje, Email: laura.mataseje@phac-aspc.gc.ca.
Anne-Catrin Uhlemann, Columbia University Irving Medical Center, New York, USA.
DATA AVAILABILITY
Raw sequencing reads were deposited to the NCBI SRA archive under BioProject PRJNA855907. See Table S1 for a list of accessions. Complete blaKPC-encoding plasmid sequences were deposited to NCBI GenBank under the accessions listed in Table S2. Updated SNV profiles used for Tn4401 typing with TETyper are found in Table S3 and updated structural profiles in are found in Table S4.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.00860-23.
Prediction of blaKPC-encoding plasmids from incomplete assemblies using MOB-recon.
Additional method details.
blaKPC-encoding isolate summary data.
Complete blaKPC-encoding plasmid summary data.
Updated Tn4401 SNV table used for TETyper analysis.
Updated Tn4401 structural table used for TETyper analysis.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Sheu C-C, Chang Y-T, Lin S-Y, Chen Y-H, Hsueh P-R. 2019. Infections caused by carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front Microbiol 10:80. doi: 10.3389/fmicb.2019.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kopotsa K, Osei Sekyere J, Mbelle NM. 2019. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann N Y Acad Sci 1457:61–91. doi: 10.1111/nyas.14223 [DOI] [PubMed] [Google Scholar]
- 4. van Duin D, Doi Y. 2017. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8:460–469. doi: 10.1080/21505594.2016.1222343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Duin D, Arias CA, Komarow L, Chen L, Hanson BM, Weston G, Cober E, Garner OB, Jacob JT, Satlin MJ, et al. 2020. Molecular and clinical epidemiology of carbapenem-resistant Enterobacteriaceae in the United States: a prospective cohort study. Lancet Infect Dis 20:731–741. doi: 10.1016/S1473-3099(19)30755-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mataseje LF, Boyd DA, Fuller J, Haldane D, Hoang L, Lefebvre B, Melano RG, Poutanen S, Van Caeseele P, Mulvey MR. 2018. Characterization of OXA-48-like carbapenemase producers in Canada, 2011–14. J Antimicrob Chemother 73:626–633. doi: 10.1093/jac/dkx462 [DOI] [PubMed] [Google Scholar]
- 7. Canadian Nosocomial Infection Surveillance Program . 2023. Healthcare-associated infections and antimicrobial resistance in Canadian acute care hospitals, 2017–2021. Can Commun Dis Rep 49:235–252. doi: 10.14745/ccdr.v49i05a09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pitout JDD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. doi: 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228–236. doi: 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- 11. Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mathers AJ, Peirano G, Pitout JDD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. David S, Cohen V, Reuter S, Sheppard AE, Giani T, Parkhill J, European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group, ESCMID Study Group for Epidemiological Markers (ESGEM), Rossolini GM, Feil EJ, Grundmann H, Aanensen DM. 2020. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc Natl Acad Sci U S A 117:25043–25054. doi: 10.1073/pnas.2003407117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tofteland S, Naseer U, Lislevand JH, Sundsfjord A, Samuelsen O. 2013. A long-term low-frequency hospital outbreak of KPC-producing Klebsiella pneumoniae involving Intergenus plasmid diffusion and a persisting environmental reservoir. PLoS One 8:e59015. doi: 10.1371/journal.pone.0059015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davies TJ, Stoesser N, Sheppard AE, Abuoun M, Fowler P, Swann J, Quan TP, Griffiths D, Vaughan A, Morgan M, Phan HTT, Jeffery KJ, Andersson M, Ellington MJ, Ekelund O, Woodford N, Mathers AJ, Bonomo RA, Crook DW, Peto TEA, Anjum MF, Walker AS. 2020. Genomic epidemiology of complex, multispecies, plasmid-borne blaKPC carbapenemase in Enterobacterales in the United Kingdom from 2009 to 2014. Antimicrob Agents Chemother 64:e02244–19. doi: 10.1128/AAC.02026-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, Khil P, Odom RT, Deming C, Park M, Thomas PJ, NISC Comparative Sequencing Program, Henderson DK, Palmore TN, Segre JA, Frank KM. 2018. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. doi: 10.1128/mBio.02011-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shropshire WC, Dinh AQ, Earley M, Komarow L, Panesso D, Rydell K, Gómez-Villegas SI, Miao H, Hill C, Chen L, Patel R, Fries BC, Abbo L, Cober E, Revolinski S, Luterbach CL, Chambers H, Fowler VG, Bonomo RA, Shelburne SA, Kreiswirth BN, van Duin D, Hanson BM, Arias CA. 2022. Accessory genomes drive independent spread of carbapenem-resistant Klebsiella pneumoniae clonal groups 258 and 307 in Houston, TX. mBio 13:e0049722. doi: 10.1128/mbio.00497-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saavedra SY, Bernal JF, Montilla-Escudero E, Arévalo SA, Prada DA, Valencia MF, Moreno J, Hidalgo AM, García-Vega ÁS, Abrudan M, Argimón S, Kekre M, Underwood A, Aanensen DM, Duarte C, Donado-Godoy P, NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance . 2021. Complexity of genomic epidemiology of carbapenem-resistant Klebsiella pneumoniae isolates in Colombia urges the reinforcement of whole genome sequencing-based surveillance programs. Clin Infect Dis 73:S290–S299. doi: 10.1093/cid/ciab777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stoesser N, Sheppard AE, Peirano G, Anson LW, Pankhurst L, Sebra R, Phan HTT, Kasarskis A, Mathers AJ, Peto TEA, Bradford P, Motyl MR, Walker AS, Crook DW, Pitout JD. 2017. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci Rep 7:5917. doi: 10.1038/s41598-017-06256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brandt C, Viehweger A, Singh A, Pletz MW, Wibberg D, Kalinowski J, Lerch S, Müller B, Makarewicz O. 2019. Assessing genetic diversity and similarity of 435 KPC-carrying plasmids. Sci Rep 9:11223. doi: 10.1038/s41598-019-47758-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 55:5370–5373. doi: 10.1128/AAC.05202-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Naas T, Cuzon G, Villegas M-V, Lartigue M-F, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sheppard AE, Stoesser N, German-Mesner I, Vegesana K, Sarah Walker A, Crook DW, Mathers AJ. 2018. TETyper: a bioinformatic pipeline for classifying variation and genetic contexts of transposable elements from short-read whole-genome sequencing data. Microb Genom 4:e000232. doi: 10.1099/mgen.0.000232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robertson J, Nash JHE. 2018. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom 4:e000206. doi: 10.1099/mgen.0.000206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmartz GP, Hartung A, Hirsch P, Kern F, Fehlmann T, Müller R, Keller A. 2022. PLSDB: advancing a comprehensive database of bacterial plasmids. Nucleic Acids Res 50:D273–D278. doi: 10.1093/nar/gkab1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robertson J, Bessonov K, Schonfeld J, Nash JHE. 2020. Universal whole-sequence-based plasmid typing and its utility to prediction of host range and epidemiological surveillance. Microb Genom 6:mgen000435. doi: 10.1099/mgen.0.000435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Redondo-Salvo S, Bartomeus-Peñalver R, Vielva L, Tagg KA, Webb HE, Fernández-López R, de la Cruz F. 2021. COPLA, a Taxonomic Classifier of Plasmids. BMC Bioinformatics 22:390. doi: 10.1186/s12859-021-04299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petkau A, Mabon P, Sieffert C, Knox NC, Cabral J, Iskander M, Iskander M, Weedmark K, Zaheer R, Katz LS, Nadon C, Reimer A, Taboada E, Beiko RG, Hsiao W, Brinkman F, Graham M, Van Domselaar G. 2017. SNVPhyl: a single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microb Genom 3:e000116. doi: 10.1099/mgen.0.000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother 54:4493–4496. doi: 10.1128/AAC.00175-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, Rojtman AD, Levi MH, Bonomo RA, Kreiswirth BN. 2014. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York hospitals. Antimicrob Agents Chemother 58:2871–2877. doi: 10.1128/AAC.00120-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Partridge SR, Ginn AN, Wiklendt AM, Ellem J, Wong JSJ, Ingram P, Guy S, Garner S, Iredell JR. 2015. Emergence of blaKPC carbapenemase genes in Australia. Int J Antimicrob Agents 45:130–136. doi: 10.1016/j.ijantimicag.2014.10.006 [DOI] [PubMed] [Google Scholar]
- 32. Arredondo-Alonso S, Willems RJ, van Schaik W, Schürch AC. 2017. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genom 3:e000128. doi: 10.1099/mgen.0.000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hopkins KL, Ellaby N, Ellington MJ, Doumith M, Mustafa N, Meunier D, Woodford N. 2022. Diversity of carbapenemase-producing Enterobacterales in England as revealed by whole-genome sequencing of isolates referred to a national reference laboratory over a 30-month period. J Med Microbiol 71. doi: 10.1099/jmm.0.001518 [DOI] [PubMed] [Google Scholar]
- 34. Lee J-A, Du S-H, Lee T-F, Huang Y-S, Liao C-H, Huang Y-T, Hsueh P-R. 2023. Rapid emergence of ceftazidime-avibactam resistance among carbapenem-resistant Enterobacterales in a tertiary-care hospital in Taiwan. J Infect 86:66–117. doi: 10.1016/j.jinf.2022.10.003 [DOI] [PubMed] [Google Scholar]
- 35. Han M, Liu C, Xie H, Zheng J, Zhang Y, Li C, Shen H, Cao X. 2023. Genomic and clinical characteristics of carbapenem-resistant Enterobacter cloacae complex isolates collected in a Chinese tertiary hospital during 2013–2021. Front Microbiol 14:1127948. doi: 10.3389/fmicb.2023.1127948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou H, Wang S, Wu Y, Dong N, Ju X, Cai C, Li R, Li Y, Liu C, Lu J, Chan E-C, Chen S, Zhang R, Shen Z. 2022. Carriage of the mcr-9 and mcr-10 genes in clinical strains of the Enterobacter cloacae complex in China: a prevalence and molecular epidemiology study. Int J Antimicrob Agents 60:106645. doi: 10.1016/j.ijantimicag.2022.106645 [DOI] [PubMed] [Google Scholar]
- 37. Huang Y-S, Chen P-Y, Chou P-C, Wang J-T. 2023. In vitro activities and Inoculum effects of cefiderocol and aztreonam-avibactam against metallo-Β-lactamase-producing Enterobacteriaceae. Microbiol Spectr 11:e0056923. doi: 10.1128/spectrum.00569-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hendrickx APA, Landman F, de Haan A, Borst D, Witteveen S, van Santen-Verheuvel MG, van der Heide HGJ, Schouls LM, Dutch CPE surveillance Study Group . 2020. Plasmid diversity among genetically related Klebsiella pneumoniae blaKPC-2 and blaKPC-3 isolates collected in the Dutch national surveillance. Sci Rep 10:16778. doi: 10.1038/s41598-020-73440-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baraniak A, Izdebski R, Fiett J, Herda M, Derde LPG, Bonten MJM, Adler A, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, MOSAR WP2, WP3, and WP5 Study Groups . 2015. KPC-like carbapenemase-producing Enterobacteriaceae colonizing patients in Europe and Israel. Antimicrob Agents Chemother 60:1912–1917. doi: 10.1128/AAC.02756-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jure MA, Castillo ME, Musa HE, López CG, Cáceres MA, Mochi SD, Bousquet AA, Genel NA, Arlet GA, Decré DC. 2019. Novel patterns in the molecular epidemiology of KPC-producing Klebsiella pneumoniae in Tucumán, Argentina. J Glob Antimicrob Resist 19:183–187. doi: 10.1016/j.jgar.2019.02.015 [DOI] [PubMed] [Google Scholar]
- 41. Findlay J, Hopkins KL, Loy R, Doumith M, Meunier D, Hill R, Pike R, Mustafa N, Livermore DM, Woodford N. 2017. OXA-48-like carbapenemases in the UK: an analysis of isolates and cases from 2007 to 2014. J Antimicrob Chemother 72:1340–1349. doi: 10.1093/jac/dkx012 [DOI] [PubMed] [Google Scholar]
- 42. León-Sampedro R, DelaFuente J, Díaz-Agero C, Crellen T, Musicha P, Rodríguez-Beltrán J, de la Vega C, Hernández-García M, López-Fresneña N, Ruiz-Garbajosa P, Cantón R, Cooper BS, San Millán Á, R-GNOSIS WP5 Study Group . 2021. Pervasive transmission of a carbapenem resistance plasmid in the gut microbiota of hospitalized patients. Nat Microbiol 6:606–616. doi: 10.1038/s41564-021-00879-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Macesic N, Hawkey J, Vezina B, Wisniewski JA, Cottingham H, Blakeway LV, Harshegyi T, Pragastis K, Badoordeen GZ, Dennison A, Spelman DW, Jenney AWJ, Peleg AY. 2023. Genomic dissection of endemic carbapenem resistance: metallo-beta-lactamase gene dissemination through clonal, plasmid and integron transfer pathways. bioRxiv. doi: 10.1101/2023.03.25.534241 [DOI] [PMC free article] [PubMed]
- 44. Andrade LN, Curiao T, Ferreira JC, Longo JM, Clímaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, Ribeiro VB, Barth AL, Paula MC, Baquero F, Cantón R, Darini A da C, Coque TM. 2011. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 55:3579–3583. doi: 10.1128/AAC.01783-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bryant KA, Van Schooneveld TC, Thapa I, Bastola D, Williams LO, Safranek TJ, Hinrichs SH, Rupp ME, Fey PD. 2013. KPC-4 Is encoded within a truncated Tn4401 in an IncL/M plasmid, pNE1280, isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob Agents Chemother 57:37–41. doi: 10.1128/AAC.01062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, Gales AC, Venezia SN, Quinn JP, Nordmann P. 2010. Worldwide diversity of Klebsiella pneumoniae that produce β-lactamase blaKPC-2 gene. Emerg Infect Dis 16:1349–1356. doi: 10.3201/eid1609.091389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gomez SA, Pasteran FG, Faccone D, Tijet N, Rapoport M, Lucero C, Lastovetska O, Albornoz E, Galas M, KPC Group, Melano RG, Corso A, Petroni A. 2011. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin Microbiol Infect 17:1520–1524. doi: 10.1111/j.1469-0691.2011.03600.x [DOI] [PubMed] [Google Scholar]
- 48. Eilertson B, Chen L, Chavda KD, Kreiswirth BN. 2017. Genomic characterization of two KPC-producing Klebsiella isolates collected in 1997 in New York City. Antimicrob Agents Chemother 61:e02458-16. doi: 10.1128/AAC.02458-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob Agents Chemother 53:1998–2004. doi: 10.1128/AAC.01355-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rada AM, De La Cadena E, Agudelo C, Capataz C, Orozco N, Pallares C, Dinh AQ, Panesso D, Ríos R, Diaz L, Correa A, Hanson BM, Villegas MV, Arias CA, Restrepo E. 2020. Dynamics of blaKPC-2 dissemination from non-CG258 Klebsiella pneumoniae to other Enterobacterales via IncN plasmids in an area of high endemicity. Antimicrob Agents Chemother 64:e01743-20. doi: 10.1128/AAC.01743-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schweizer C, Bischoff P, Bender J, Kola A, Gastmeier P, Hummel M, Klefisch F-R, Schoenrath F, Frühauf A, Pfeifer Y. 2019. Plasmid-mediated transmission of KPC-2 carbapenemase in Enterobacteriaceae in critically Ill patients. Front Microbiol 10:276. doi: 10.3389/fmicb.2019.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hazen TH, Mettus R, McElheny CL, Bowler SL, Nagaraj S, Doi Y, Rasko DA. 2018. Diversity among blaKPC-containing plasmids in Escherichia coli and other bacterial species isolated from the same patients. Sci Rep 8:10291. doi: 10.1038/s41598-018-28085-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tijet N, Muller MP, Matukas LM, Khan A, Patel SN, Melano RG. 2016. Lateral dissemination and inter-patient transmission of blaKPC-3: role of a conjugative plasmid in spreading carbapenem resistance. J Antimicrob Chemother 71:344–347. doi: 10.1093/jac/dkv356 [DOI] [PubMed] [Google Scholar]
- 54. Mataseje LF, Abdesselam K, Vachon J, Mitchel R, Bryce E, Roscoe D, Boyd DA, Embree J, Katz K, Kibsey P, Simor AE, Taylor G, Turgeon N, Langley J, Gravel D, Amaratunga K, Mulvey MR. 2016. Results from the Canadian nosocomial infection surveillance program on carbapenemase-producing Enterobacteriaceae, 2010 to 2014. Antimicrob Agents Chemother 60:6787–6794. doi: 10.1128/AAC.01359-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stohr J, Verweij JJ, Buiting AGM, Rossen JWA, Kluytmans J. 2020. Within-patient plasmid dynamics in Klebsiella pneumoniae during an outbreak of a carbapenemase-producing Klebsiella pneumoniae. PLoS One 15:e0233313. doi: 10.1371/journal.pone.0233313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Prussing C, Snavely EA, Singh N, Lapierre P, Lasek-Nesselquist E, Mitchell K, Haas W, Owsiak R, Nazarian E, Musser KA. 2020. Nanopore MinION sequencing reveals possible transfer of blaKPC-2 plasmid across bacterial species in two healthcare facilities. Front Microbiol 11:2007. doi: 10.3389/fmicb.2020.02007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martin J, Phan HTT, Findlay J, Stoesser N, Pankhurst L, Navickaite I, De Maio N, Eyre DW, Toogood G, Orsi NM, Kirby A, Young N, Turton JF, Hill RLR, Hopkins KL, Woodford N, Peto TEA, Walker AS, Crook DW, Wilcox MH. 2017. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J Antimicrob Chemother 72:3025–3034. doi: 10.1093/jac/dkx264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Neffe L, Abendroth L, Bautsch W, Häussler S, Tomasch J. 2022. High plasmidome diversity of extended-spectrum beta-lactam-resistant Escherichia coli isolates collected during one year in one community hospital. Genomics 114:110368. doi: 10.1016/j.ygeno.2022.110368 [DOI] [PubMed] [Google Scholar]
- 59. Barry KE, Wailan AM, Sheppard AE, Crook D, Vegesana K, Stoesser N, Parikh HI, Sebra R, Mathers AJ. 2019. Don’t overlook the little guy: an evaluation of the frequency of small plasmids co-conjugating with larger carbapenemase gene containing plasmids. Plasmid 103:1–8. doi: 10.1016/j.plasmid.2019.03.005 [DOI] [PubMed] [Google Scholar]
- 60. Viehweger A, Blumenscheit C, Lippmann N, Wyres KL, Brandt C, Hans JB, Hölzer M, Irber L, Gatermann S, Lübbert C, Pletz MW, Holt KE, König B. 2021. Context-aware genomic surveillance reveals hidden transmission of a carbapenemase-producing Klebsiella pneumoniae. Microb Genom 7:000741. doi: 10.1099/mgen.0.000741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, de Man T, Rasheed JK, Engelthaler DM, Keim P, Limbago BM. 2015. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 10:e0133727. doi: 10.1371/journal.pone.0133727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pallecchi L, Riccobono E, Mantella A, Fernandez C, Bartalesi F, Rodriguez H, Gotuzzo E, Bartoloni A, Rossolini GM. 2011. Small qnrB-harbouring ColE-like plasmids widespread in commensal enterobacteria from a remote Amazonas population not exposed to antibiotics. J Antimicrob Chemother 66:1176–1178. doi: 10.1093/jac/dkr026 [DOI] [PubMed] [Google Scholar]
- 63. Sugita K, Aoki K, Komori K, Nagasawa T, Ishii Y, Iwata S, Tateda K. 2021. Molecular analysis of blaKPC-2-harboring plasmids: Tn4401a interplasmid transposition and Tn4401a-carrying ColRNAI plasmid mobilization from Klebsiella pneumoniae to Citrobacter europaeus and Morganella morganii in a single patient. mSphere 6:e0085021. doi: 10.1128/mSphere.00850-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moran RA, Hall RM. 2018. Evolution of regions containing antibiotic resistance genes in FII-2-FIB-1 ColV-Colla virulence plasmids. Microb Drug Resist 24:411–421. doi: 10.1089/mdr.2017.0177 [DOI] [PubMed] [Google Scholar]
- 65. Robertson J, Schonfeld J, Bessonov K, Bastedo P, Nash JHE. 2023. A global survey of Salmonella plasmids and their associations with antimicrobial resistance. Microb Genom 9:mgen001002. doi: 10.1099/mgen.0.001002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, Anson LW, Giess A, Pankhurst LJ, Vaughan A, Grim CJ, Cox HL, Yeh AJ, Modernising Medical Microbiology (MMM) Informatics Group, Sifri CD, Walker AS, Peto TE, Crook DW, Mathers AJ. 2016. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 60:3767–3778. doi: 10.1128/AAC.00464-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Orlek A, Stoesser N, Anjum MF, Doumith M, Ellington MJ, Peto T, Crook D, Woodford N, Walker AS, Phan H, Sheppard AE. 2017. Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front Microbiol 8:182. doi: 10.3389/fmicb.2017.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. doi: 10.1038/s41467-021-24448-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brehony C, McGrath E, Brennan W, Tuohy A, Whyte T, Brisse S, Maiden M, Jolley K, Morris D, Cormican M. 2019. An MLST approach to support tracking of plasmids carrying OXA-48-like carbapenemase. J Antimicrob Chemother 74:1856–1862. doi: 10.1093/jac/dkz136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pierce NT, Irber L, Reiter T, Brooks P, Brown CT. 2019. Large-scale sequence comparisons with sourmash. F1000Res 8:1006. doi: 10.12688/f1000research.19675.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Giménez M, Ferrés I, Iraola G. 2022. Improved detection and classification of plasmids from circularized and fragmented assemblies. bioRxiv. doi: 10.1101/2022.08.04.502827 [DOI]
- 72. Pradier L, Tissot T, Fiston-Lavier A-S, Bedhomme S. 2021. PlasForest: a homology-based random forest classifier for plasmid detection in genomic datasets. BMC Bioinformatics 22:349. doi: 10.1186/s12859-021-04270-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van Domselaar G. 2023. RefSeq Masher. National Microbiology laboratory. https://github.com/phac-nml/refseq_masher. [Google Scholar]
- 75. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bharat A, Petkau A, Avery BP, Chen JC, Folster JP, Carson CA, Kearney A, Nadon C, Mabon P, Thiessen J, Alexander DC, Allen V, El Bailey S, Bekal S, German GJ, Haldane D, Hoang L, Chui L, Minion J, Zahariadis G, Domselaar GV, Reid-Smith RJ, Mulvey MR. 2022. Correlation between phenotypic and in silico detection of antimicrobial resistance in Salmonella Enterica in Canada using Staramr. Microorganisms 10:292. doi: 10.3390/microorganisms10020292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seemann T. 2023. mlst. https://github.com/tseemann/mlst.
- 79. Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cheruvanky A, Stoesser N, Sheppard AE, Crook DW, Hoffman PS, Weddle E, Carroll J, Sifri CD, Chai W, Barry K, Ramakrishnan G, Mathers AJ. 2017. Enhanced Klebsiella pneumoniae carbapenemase expression from a novel Tn4401 deletion. Antimicrob Agents Chemother 61:e00025-17. doi: 10.1128/AAC.00025-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Araújo BF, Royer S, Campos PA, Ferreira ML, Gonçalves IR, Machado LG, Lincopan N, Fernandes MR, Cerdeira LT, Batistão D da F, Gontijo-Filho PP, Ribas RM. 2018. Insights into a novel Tn4401 deletion (Tn4401i) in a multidrug-resistant Klebsiella pneumoniae clinical strain belonging to the high-risk clonal group 258 producing KPC-2. Int J Antimicrob Agents 52:525–527. doi: 10.1016/j.ijantimicag.2018.08.011 [DOI] [PubMed] [Google Scholar]
- 84. Chmelnitsky I, Shklyar M, Leavitt A, Sadovsky E, Navon-Venezia S, Ben Dalak M, Edgar R, Carmeli Y. 2014. Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn Microbiol Infect Dis 79:255–260. doi: 10.1016/j.diagmicrobio.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 85. Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, Gladstone RA, Lo S, Beaudoin C, Floto RA, Frost SDW, Corander J, Bentley SD. 2020. Producing polished prokaryotic pangenomes with the panaroo pipeline. Genome Biol 21:180. doi: 10.1186/s13059-020-02090-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prediction of blaKPC-encoding plasmids from incomplete assemblies using MOB-recon.
Additional method details.
blaKPC-encoding isolate summary data.
Complete blaKPC-encoding plasmid summary data.
Updated Tn4401 SNV table used for TETyper analysis.
Updated Tn4401 structural table used for TETyper analysis.
Data Availability Statement
Raw sequencing reads were deposited to the NCBI SRA archive under BioProject PRJNA855907. See Table S1 for a list of accessions. Complete blaKPC-encoding plasmid sequences were deposited to NCBI GenBank under the accessions listed in Table S2. Updated SNV profiles used for Tn4401 typing with TETyper are found in Table S3 and updated structural profiles in are found in Table S4.