Abstract
Objective
This study is to investigate the role of gut dysbiosis in triggering inflammation in the brain and its contribution to Alzheimer’s disease (AD) pathogenesis.
Design
We analysed the gut microbiota composition of 3×Tg mice in an age-dependent manner. We generated germ-free 3×Tg mice and recolonisation of germ-free 3×Tg mice with fecal samples from both patients with AD and age-matched healthy donors.
Results
Microbial 16S rRNA sequencing revealed Bacteroides enrichment. We found a prominent reduction of cerebral amyloid-β plaques and neurofibrillary tangles pathology in germ-free 3×Tg mice as compared with specific-pathogen-free mice. And hippocampal RNAseq showed that inflammatory pathway and insulin/IGF-1 signalling in 3×Tg mice brain are aberrantly altered in the absence of gut microbiota. Poly-unsaturated fatty acid metabolites identified by metabolomic analysis, and their oxidative enzymes were selectively elevated, corresponding with microglia activation and inflammation. AD patients’ gut microbiome exacerbated AD pathologies in 3×Tg mice, associated with C/EBPβ/asparagine endopeptidase pathway activation and cognitive dysfunctions compared with healthy donors’ microbiota transplants.
Conclusions
These findings support that a complex gut microbiome is required for behavioural defects, microglia activation and AD pathologies, the gut microbiome contributes to pathologies in an AD mouse model and that dysbiosis of the human microbiome might be a risk factor for AD.
INTRODUCTION
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder that is characterised pathologically by the presence of extracellular with amyloid-β (Aβ) peptides and neurofibrillary tangles (NFT), with hyperphosphorylated and aggregated tau as major components in the brain. Chronic inflammation plays a pivotal role in AD pathogenesis, and neuroinflammation in AD brains is activated by the central stimuli, such as senile plaques and NFT1 and peripheral stimuli including endotoxins.2 Activated microglia and neuroinflammation are highly correlated with cognitive decline in patients with AD.3 Moreover, polymorphisms within genes critical for innate immune responses demonstrate an elevated risk of late-onset AD.4
The gut microbiome is an important environmental cue for microglial function in immune and neuronal responses throughout the host’s life span. The gut–brain axis, composed of immune, vagus nerve and neuroendocrine pathways,5 regulates host cognitive behaviours.6 Gut microbiota imbalance can lead to increased permeability of the intestinal epithelial barrier with the release of proinflammatory cytokines and promotion of a neuroinflammatory response.7 Patients with cognitive impairment and brain amyloidosis exhibit altered gut microbiome composition, significantly associated with proinflammatory cytokine gene profiles in these subjects.8 The abundance of Proteobacteria is highly enriched in the faeces of patients with AD. In contrast, the relative proportion of phylum Firmicutes is significantly reduced, which influences the central nervous system (CNS) immunity and neuroendocrine.9 Other studies show decreased Firmicutes, increased Bacteroidetes, and decreased Bifidobacterium in AD subjects.10 11 Furthermore, the AD patient microbiome, associated with dysregulation of the anti-inflammatory P-glycoprotein pathway, contains a lower abundance of butyrate synthesising bacteria, and higher abundances of predicted proinflammatory taxa.12 In addition to human studies, altered microbiomes have also been reported in various AD mouse models.13 14 Recently, we reported that gut dysbiosis contributes to amyloid pathology in an AD mouse model.15 Noticeably, germ-free (GF) APP/PS1 Tg mice display reduced Aβ deposition compared with their specific pathogen-free (SPF) counterparts. Colonisation of GF mice with the conventional mice’s microbiota increases Aβ pathology.16
The poly-unsaturated fatty acids (PUFAs) are categorised into two classes, omega-3 and omega-6 fatty acids. Alpha-linolenic acid (omega-3) and linoleic acid (omega-6) are parental and essential fatty acids. They are metabolically converted into long chain UFAs with arachidonic acid (AA) from linoleic acid and docosahexanenoic acid and eicosapentaenoic acid from linolenic acid. Oxidative AA metabolism is a major hallmark of neuroinflammation.17 Following its release from the cell membrane by activated phospholipase A2, AA is metabolised via cyclooxygenases (COX-1, 2) and 5-lipoxygenase (5-LOX) into prostaglandins (PGs) and leukotrienes (LTs), respectively. These bioactive lipids are potent mediators of inflammation. The AA metabolic pathway is not only the core network of inflammation, but also the main cause of working memory impairment leading to AD pathogenesis.18 19 For instance, AA inhibits the muscarinic acetylcholine receptors in the AD brains, which aggravates cognitive dysfunction.20 AA metabolic level is generally elevated in the AD brains21 and in hAPP-J20 AD mice.22 COX-2 expressed by hippocampal and cortical neurons and COX-1, found in microglia, regulate neuroplasticity producing PGs.23 Notably, PGE2 and LTB4 (leukotriene B4) as intermediates in this pathway are greatly enriched in human AD brains, consistent with the previous report that AA metabolite structural features affect the production of Aβ.24 PGE2 regulates Aβ deposition and Aβ release, enhancing secretion of Aβ peptides.25 Further, 5-LOX regulates Aβ levels in the brain by influencing γ-secretase.26 Accordingly, knockout of 5-LOX rescues synaptic dysfunction and improves memory.27 Conversely, 5-LOX overexpression worsens memory, and enhances amyloid and tau brain pathologies.28
Recently, we reported that asparagine endopeptidase (AEP) simultaneously cleaves both APP and tau and stimulates Aβ production and NFT formation. AEP cleaves APP N585 and tau N368 in AD brains, and inactivation of AEP substantially diminishes AD pathologies, resulting in restoration of cognitive functions.29 30 Moreover, C/EBPβ, an Aβ and proinflammatory cytokines-activated transcription factor, regulates AEP expression in an age-dependent manner.31 Interestingly, C/EBPβ also binds the inflammatory cytokines promoters and escalates their upregulation,32 and C/EBPβ in microglia exacerbates tau-mediated AD pathology.33 Most recently, we reported that C/EBPβ/AEP signalling is age-dependently activated in 5xFAD mice, which is modulated by gut microbiota. Antibiotics diminish this signalling and attenuate amyloidogenic processes, improving cognitive functions.15 Gut-derived Aβ and tau N368 fibrils spread into the brain along the vagus nerve, triggering AD pathologies in the brain.34 In order to assess the role of the microbiota in AD aetiology, we sought to determine the functional contributions by the gut microbiota to pathologies and behaviours associated with AD. Herein, we report that colonisation of GF 3×Tg mice with faecal microbiota from individuals with AD is sufficient to promote cognitive defects and facilitate AD pathologies, relative to those with healthy control (HC) microbiota. We identify specific microbial metabolites that are sufficient to promote disease symptoms, through fatty acid-mediated inflammatory pathways. Hence, these results suggest that gut microbes may play a critical and functional role in AD pathogenesis.
RESULTS
Gut microbes stimulate AD pathologies and cognitive deficits and microglia activation in 3×Tg mice
We recently reported that the microbiota of aged 3×Tg mice accelerate AD pathology in young 3×Tg mice, accompanied by active C/EBPβ/AEP signalling in the brain,15 indicating that age-dependent microbiome changes affect AD pathology. Therefore, the longitudinal study of microbiome composition is necessary for discovering changes in microbial communities that affect the host. To identify temporal signatures in the microbiota that promote physiologic or pathophysiologic responses in an AD mouse model, we utilised the 3×Tg murine model of AD, and generated a 17-month time series of 16S rRNA gene profiles by high throughput sequencing analysis. The microbiome analysis revealed a dramatic disequilibrium in the microbial composition of the 4, 8, 12 and 17-month-old 3×Tg mice. In addition, a principal coordinate plot (PCoA) demonstrated that the microbiota of 3×Tg mice clustered distinctly from that of young wild-type mice (online supplemental figure 1A,B). At the phylum level, the relative abundances of Proteobacteria were escalated in 4, 12 and 17-month-old 3×Tg mice in comparison with 4-month-old WT (online supplemental figure 1B–D). On the other hand, Firmicutes and Cyanobacteria’s relative abundances gradually decreased, and, in contrast, Bacteroidetes were dramatically increased in 8 and 17-month-old 3xTg mice compared with with 4-month-old 3×Tg mice. A depletion of phyla Firmicutes and an increase in Bacteroidetes are associated with augmented inflammation and different diseases. In addition, mucin degrading and inflammation-inducing Ruminococcus species demonstrated increased relative frequencies in the aged 3×Tg mice (online supplemental figure 2A–C). In brief, the microbiome analysis demonstrates that the aged 3×Tg mice harbour an altered microbiota, which is characterised by depleted community of beneficial anti-inflammatory bacteria followed by the enrichment of proinflammatory pathobionts of the intestine (online supplemental figure 1C,G).
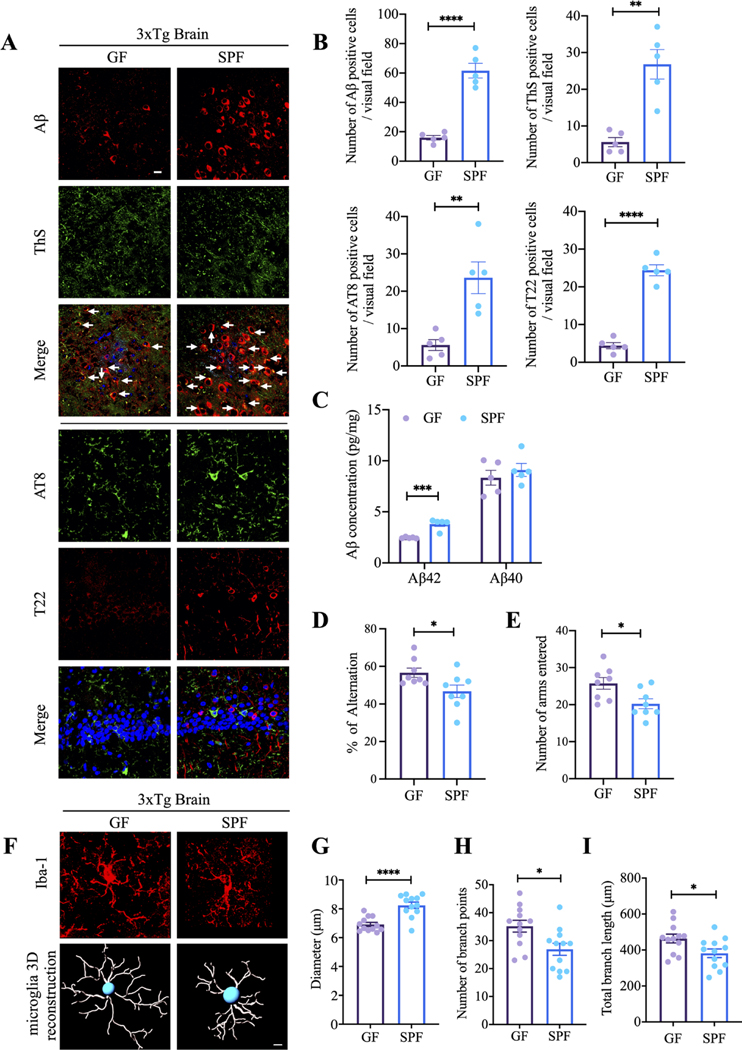
To address the gut microbiota’s role in AD pathogenesis, we rederived GF the 3×Tg AD mouse model. On average, GF mice body weights were lighter than age-matched SPF littermates. Moreover, GF mice exhibited enlarged caeca with increased weights and dark-coloured cecal contents as a consequence of absent gut microbiomes (online supplemental figure 3A,B), though their GI tract length remained comparable to the counterpart of SPF mice (online supplemental figure 3C,D). The bacteria-free status from the faecal samples from GF mice was further validated by bacterial cultures, which displayed no demonstrable bacteria (online supplemental figure 3E,F). Compared with SPF 3×Tg mice, deposited Aβ aggregates in the cortex and ThS (Thioflavin S) immunofluorescent (IF) co-staining signals significantly decreased in GF mice. Moreover, hyperphosphorylated tau (AT8) in the hippocampus and fibrillar tau inclusions, validated with specific T22 monoclonal antibody, were also greatly decreased in GF mice versus SPF mice (figure 1A,B). Quantification revealed that Aβ42, but not Aβ40, was diminished in GF mice as compared with SPF mice (figure 1C). These observations indicate that AD pathologies are alleviated when the gut microbiota are depleted.
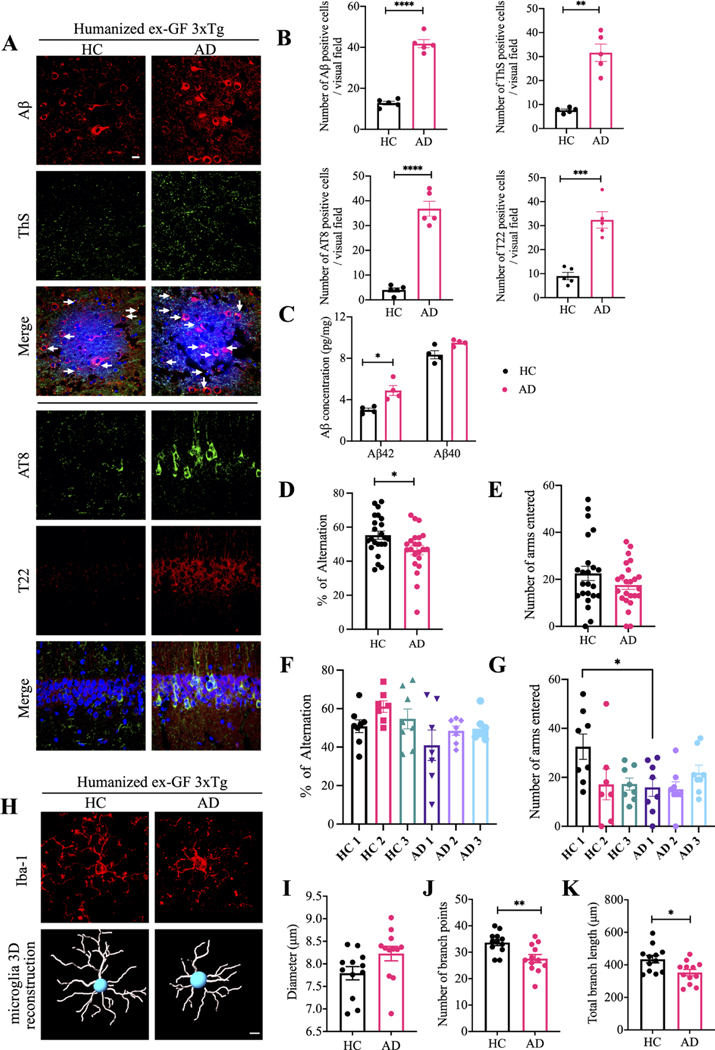
Figure 1.
Germ-free 3xTg mice display reduced AD pathologies and improved cognitive functions compared with SPF 3xTg mice.
(A) Immunofluorescent staining of Aβ (red) and ThS (green) in frontal cortex region of brains, AT8 (green) and T22 (red) in hippocampus CA1 region of brains from germ-free 3xTg mice and SPF mice. Scale bar: 20 μm (B) quantitative analysis of Aβ positive cells, ThS positive cells, AT8 positive cells and T22 positive cells, respectively. The density of Aβ, ThS, AT8 and T22 positive cells were significantly increased in spf mice brain. (n=5 in each group, data are shown as mean±SEM. **P<0.01, ****p<0.0001 compared with control, unpaired t-tests.). (C) Aβ40 and Aβ42 concentrations in the cortex of germ-free 3xTg mice and SPF 3xTg mice were measured using human aβ40 and Aβ42 ELISA kit. the concentration of Aβ42 not aβ40 was significantly increased in spf 3xTg mice cortex compared with germ-free 3xTg mice cortex. (n=5 in each group, data are shown as mean±SEM ***p<0.001 compared with control, multiple unpaired T tests). (D, E) Y-maze behavioural tests. Spontaneous alternation (%) (D), number of arms entered (E); n=8 in each group, data are shown as mean±SEM. *P<0.05 compared with control, unpaired T tests. (F) Representative images of immunofluorescent staining of Iba-1 (red) in cortex (upper panel) and 3D reconstruction of Iba-1-stained microglia (lower panel) residing in the cortex of germ-free 3xTg mice and SPF 3xTg mice. (G–I) Quantitative analysis of diameter, number of branch points, and total branch length of microglia residing in the cortex. Data represent the mean±SEM; representative data of 12 samples; *p<0.05, ****p<0.0001 compared with control, unpaired t-tests. Aβ, amyloid-β; GF, germ-free; SPF, specific-pathogen-free.
Though the levels of BDNF, an essential neurotrophine for synaptogenesis and dendritic arborisation, in the brains from both mice were similar (online supplemental figure 3G), Golgi staining showed that the dendritic spines were highly elevated in the hippocampal neurons from GF mice as compared with SPF mice (online supplemental figure 3H,I). Concurrently, behaviour in the Y-maze showed that spatial memory functions were significantly improved for GF mice versus SPF mice (figure 1D,E). Microglia maturation and activation are associated with chronic neuroinflammation in AD brains. Accordingly, Iba-1 staining demonstrated significantly longer processes and increased numbers of segments, branching and terminal points in the microglia cells in the cortex from GF mice. Semiautomatic quantitative morphometric three-dimensional measurements of microglia revealed that these microglial morphologies were markedly changed in GF mice compared with SPF mice (figure 1F–I). Thus, gut microbiota facilitates AD pathologies, cognitive deficits and microglia activation.
Gut microbes activate C/EBPβ/AEP pathway, elevating proinflammatory enzymes implicated in PUFA metabolism
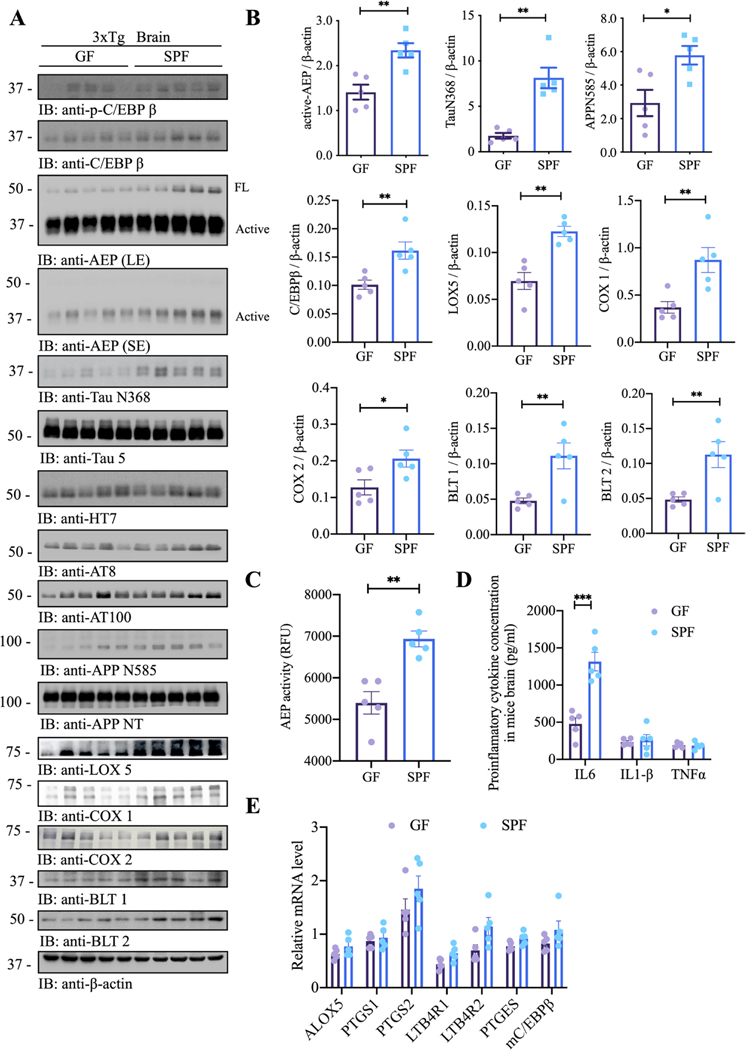
Our recent studies support that C/EBPβ/AEP signalling tempo-spatially regulates AD pathologies.31 35 To assess whether an intact gut microbiome was necessary to stimulate this pathway, contributing to AD pathogenesis, we conducted immunoblotting analysis and found that C/EBPβ/AEP signalling was attenuated in the brains of GF 3xTg mice as compared with those of SFP mice. Subsequently, AEP-truncated tau N368 and APP N585 fragments were reduced, associated with lower p-tau AT8 and AT100 activities. Remarkably, Lox5, Cox1 and Cox2 levels were evidently lessened in GF 3×Tg mice versus SPF mice (figure 2A,B), fitting with previous findings that AA metabolic pathway is not only the core network of inflammation but also the main cause of working memory impairment leading to AD pathogenesis.18 19 As expected, protease assay showed that AEP enzymatic activities in GF mice were lower than SPF mice, consistent with reduced active AEP levels (figure 2C). Since C/EBPβ is a primary transcription factor for IL-6, accordingly, IL-6 concentrations in GF mice were significantly decreased as compared with SPF mice (figure 2D). qRT-PCR revealed that CEBPβ targets/AA pathway genes were slightly reduced in GF mice in comparison to SPF mice (figure 2E), corresponding with the RNAseq data. However, the qRT-PCR data are no statistically significant, which might be due to the high variation in quantitative PCR (qPCR) analysis.
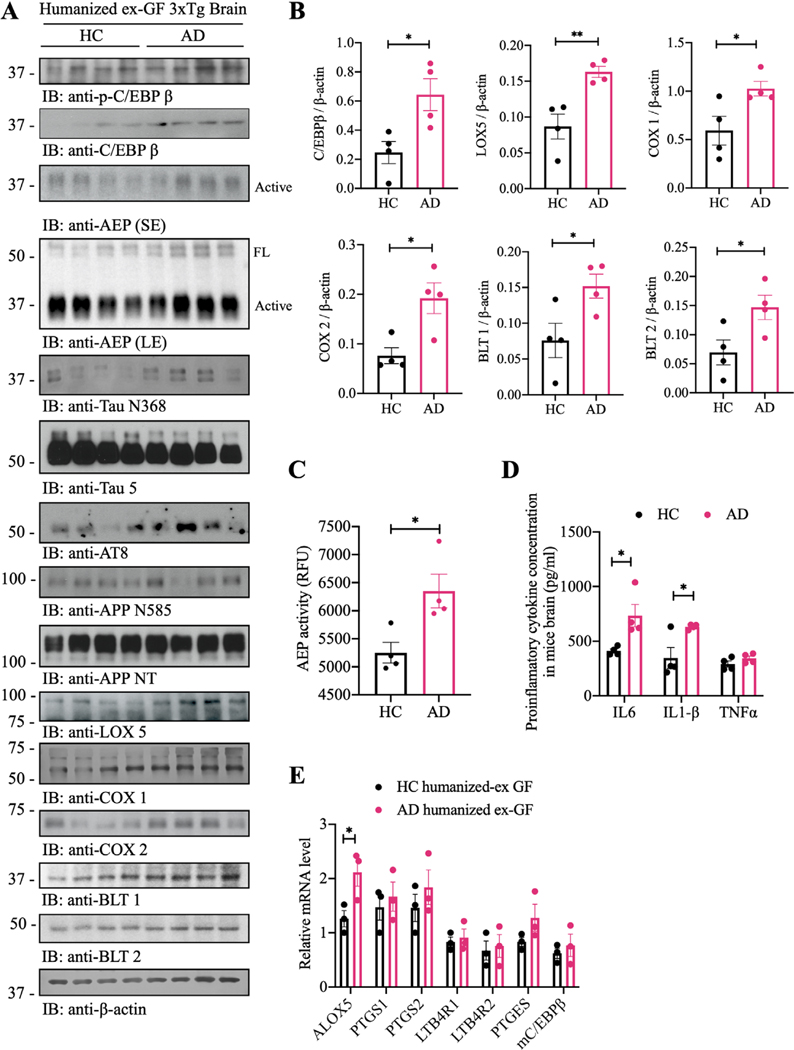
Figure 2.
Germ-free (GF) 3xTg mice demonstrate attenuated C/EBPβ/AEP pathway and AA-associated inflammation. (A) immunoblot showing p-C/EBPβ, C/EBPβ, AEP, APP and tau expression and processing, as well as arachidonic acid metabolism in mouse brains of germ-free 3xTg mice and SPF 3xTg mice. (B) Quantitative analysis of immunoblot. The bands of active-AEP, TauN368, APPN585, C/EBPβ, LOX5, Cox1, COX2, BLT1 and BLT2 were measured with image J and normalised with β-actin. n=5 in each group, data are shown as mean±SEM, *p<0.05, **p<0.01 compared with control, unpaired t-tests. (C) AEP activity assay in the brain lysates from germ-free 3xTg mice and SPF 3xTg mice. SPF 3xTg mice showed escalation in AEP activities compared with germ-free 3xTg mice. Data represent the mean±SEM; representative data of five samples; **p<0.01 compared with control, unpaired t-tests. (D) proinflammatory cytokine IL1-β, IL-6 and TNFα concentrations in the brain lysates from germ-free 3xTg mice and SPF 3xTg mice, respectively. Data represent the mean±SEM; representative data of five samples; ***p<0.001 compared with control, multiple unpaired t-tests. (E) Quantitative RT-PCR analysis of the brain samples from GF mice versus spf mice comparing C/EBPb targeted AA pathway genes. AA, arachidonic acid; AEP, asparagine endopeptidase; COX, cyclooxygenases; SPF, specific-pathogen-free.
IF co-staining of brain sections revealed reduction of C/EBPβ upregulation, AEP activation, Aβ accumulation, Tau N368 fragmentation and p-Tau (AT8) in the hippocampus and the cortex of GF mice vs SPF mice (oonline supplemental figure 4A–D, hippocampus: A and D; cortex: B and C), in alignment with decreased C/EBPβ/AEP signalling and attenuated APP N585 and Tau N368 fragmentation in GF mice. Moreover, IF activities of 5-Lox, Cox1 and Cox2 in the brains of GF mice were manifestly diminished, tightly correlated with lessened C/EBPβ signals (online supplemental figure 4E–H, cortex: E, F and G). Thus, C/EBPβ/AEP signalling, and AA-associated inflammatory enzymes were diminished in GF mice vs SPF mice, suggesting that gut microbiota regulates these effectors’ expression via activating C/EBPβ/AEP pathway. These findings are consistent with previous reports that C/EBPβ acts as a transcription factor for these AA-associated inflammatory enzymes.36 Immunohistochemistry (IHC) staining further validated that Iba-1 and AEP both were decreased in the brains of GF mice as compared with SPF mice (online supplemental figure 4I), consistent with Iba1 and AEP IF co-staining in the cortex. Hence, GF 3×Tg mice demonstrate attenuated C/EBPβ/AEP pathway and AA-associated inflammation.
Gut microbiota elevates inflammatory metabolites and alters inflammatory and insulin/IGF-1 pathways in the brain
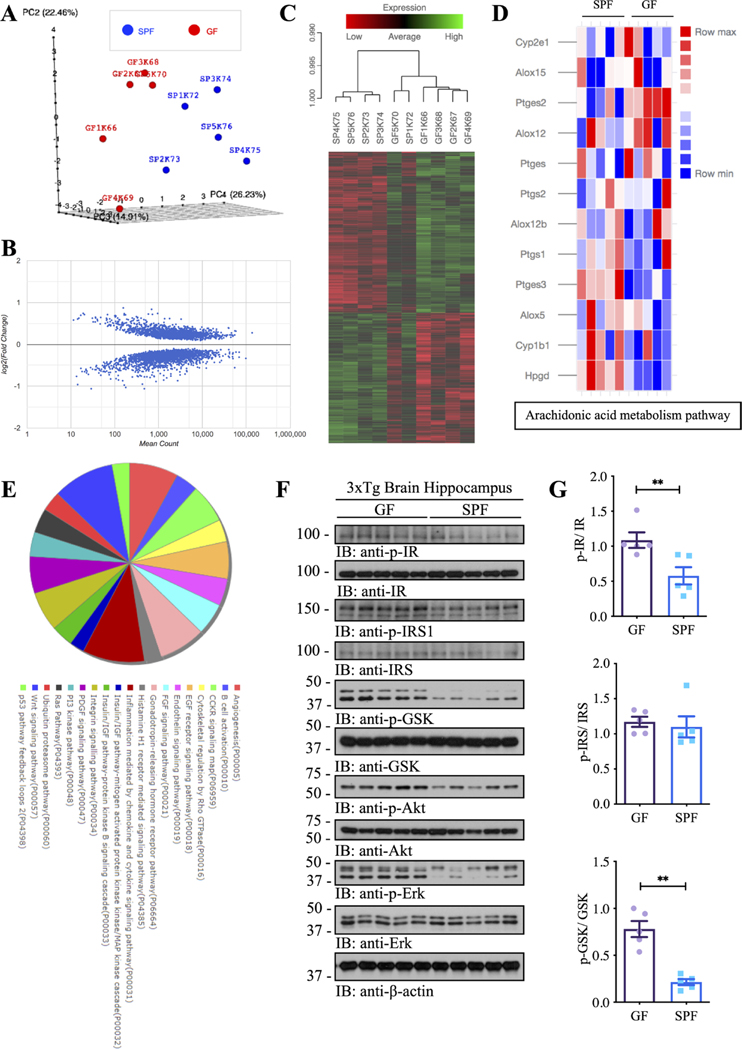
To evaluate the global impact of the gut microbiome on AD-associated transcriptional response, we performed unbiased RNA-sequencing using the hippocampal samples from GF and SPF 3×Tg mice and measured the genome-wide mRNA expression profiles by quantitative deep sequencing of the RNA transcripts (figure 3 and online supplemental figure 5A,B). We found marked differences in the mRNA profiles and overall gene expression pattern between SPF and GF animals. Assessment of the primary sources of variance in the dataset by principal component analysis (PCA) revealed strong separation (figure 3A) of GF and SPF 3×Tg mice. We observed 1872 differential genes in SPF 3×Tg mice compared with GF 3×Tg counterparts (figure 3B). Among the differentially expressed genes, we observed in the hippocampus from SPF 3×Tg mice downregulated expression of apolipoprotein E compared with the GF 3×Tg mice. Next, the hippocampal genes differentially expressed between GF and SPF 3×Tg mice were used to generate heatmaps by cluster analysis of functional mRNAs (figure 3C). Strikingly, genes involved in AA metabolism pathways, including Hpgd, Ptges3, Ptges2, and Cyp1b1, were differentially expressed in SPF 3xTg mice in comparison with GF 3xTg mice (upregulated: Hpgd, Ptges3, and Cyp1b1; downregulated: Ptges2) (figure 3D), fitting with our observations that numerous AA-associated inflammatory enzymes are clearly reduced in GF mice. In addition to the profoundly changed proinflammatory genes in the AA metabolism pathway, interestingly, RNAseq revealed that insulin signalling, and its associated downstream pathways were abnormally altered in SPF mice as compared with GF mice (figure 3E). Therefore, we examined hippocampal insulin signalling by immunoblotting, and found that both p-insulin receptor (IR) and p-IRS (IR substrate) and their downstream effectors including p-Akt and p-MAPK and p-GSK activities were strongly escalated in GF mice as compared with SPF mice (figure 3F). These data are consistent with previous reports that AD patients show progressive brain insulin resistance and insulin deficiency.37 38
Figure 3.
Gutmicrobiota impacts the transcriptome atlas of mRNA expression in the hippocampal regions of germ-free and SPF 3xTg mouse model of AD. (A) Plot showing sample-to-sample distances in a principal components analysis (PCA). (B) Scatter plot showing log2 (fold change) against normalised mean gene count. (C) Cluster analysis of functional mRNAs in GF and SPF, and data are presented as a heat MAP (p<0.05). Each row represents the relative levels of expression of a single gene across all mice; each column represents the expression levels for a single mouse. The colours red and green denote low and high expression, respectively. (D) Heatmap showing differential genes of Arachidonic acid metabolism pathways between GF and SPF. (E) KEGG and Panther pathway analysis of differential genes. (F) Immunoblot blot showing insulin/IGF-1 signalling pathways in the hippocampus of germ-free 3xTg mice and SPF 3xTg mice. (G) Quantitative analysis of immunoblot. The bands of p-IR, p-IRS and p-GSK were measured with image J and normalised with IR, IRS and GSK, respectively. N=5 in each group, data are shown as mean±SEM, **p<0.01 compared with control, unpaired t-tests. AD, Alzheimer’s disease; GF, germ-free; IR, insulin receptors; SPF, specific-pathogen-free.
Next, we performed enrichment analyses of differentially expressed genes in the hippocampus using a curated collection of public gene sets (GO, KEGG and PANTHER) (online supplemental figure 5C,D), and identified alterations in genes involved in apoptotic, serotonin and dopamine, GABAnergic, glutamate, and norepinephrine pathways (online supplemental figure 5E–I). As shown, proapoptotic genes including Tnf, Apaf1, Fasl, etc were increased and anti-apoptotic gene Bcl2l1 was decreased in SPF mice, although proapoptotic genes including Bax, Bad, Casp3,etc were also decreased in SPF mice, indicating that different pathways-mediated cell death effectors were greatly altered between SPF and GF mice and the cells in GF 3×Tg brains might be more resistant against apoptosis (online supplemental figure 5E). Slc6a4 gene that encodes an integral membrane protein transporting the neurotransmitter serotonin was decreased in SPF mice, and Htr3b, Htr4 genes, which encode the receptors for serotonin were also decreased in SPF mice, indicating the potential for increased serotonin signalling in the hippocampus of GF mice (online supplemental figure 5F). Ankk1 gene is closely linked to Drd2 gene, and both were decreased in SPF mice, while Drd2, Drd3, Drd4 and Drd5 that encode the dopamine receptors were also decreased in SPF mice, suggesting more dopamine receptors were expressed in GF mice (online supplemental figure 5F). Gad1 encodes glutamic acid decarboxylase that is responsible for catalysing the production of γ-aminobutyric acid (GABA) from L-glutamic acid; Aldh9a1 gene encodes the enzyme oxidising γ-amino butyraldehyde to GABA; Gabarap gene encodes GABA(A) receptor-associated protein. All of these were decreased in SPF mice, while Abat gene that encodes 4-aminobutyrate aminotransferase responsible for catabolism of GABA was increased in SPF mice, indicating that more GABA is produced and uptaken in GF 3×Tg mice hippocampus without degradation, and more GABA receptors synthesised in GF mice compared with SPF mice (online supplemental figure 5G); Gls and Gls2 genes encode glutaminases catalysing the hydrolysis of glutamine to glutamate and ammonia; Glul gene encodes the protein that synthesises glutamine from glutamate; Grin1, Grin2a, Grin2b genes encode the subunits of N-methyl-D-aspartate receptors; All of the above genes were increased in the hippocampus of SPF 3×Tg mice, while less glutamate is produced in GF mouse brains, accompanied with higher glutamate degradation and less glutamate receptors synthesis, compared with SPF mice (online supplemental figures 5H and 7H). Comt gene encodes Catechol-O-methyltransferase; Dbh gene encodes the oxidoreductase catalysing the conversion of dopamine to norepinephrine, and they were decreased in SPF vs GF, while Adra1a and Adra2a genes that encode alpha-1A-adrenergic and alpha-2A-adrenergic receptors were increased in SPF mice, indicating that less norepinephrine is produced in GF mouse brains (online supplemental figure 5I). Two keystone genes for each pathway were validated by qRT-PCR, and results were consistent with RNA-Seq findings (online supplemental figure 5J). NeuN and TUNEL co-staining showed that the apoptosis in the hippocampal neurons in SPF mice was greatly increased (online supplemental figure 5K). Together, these data indicate that resident gut microbiota impacts the transcriptomic landscape of mRNA expression in the hippocampal regions of GF and SPF 3xTg mouse model of AD.
Short chain fatty acids trigger C/EBPβ/AEP activation and cognitive deficits in GF 3×Tg mice and inflammation, exacerbated by prostaglandin E2–1-glyceryl ester
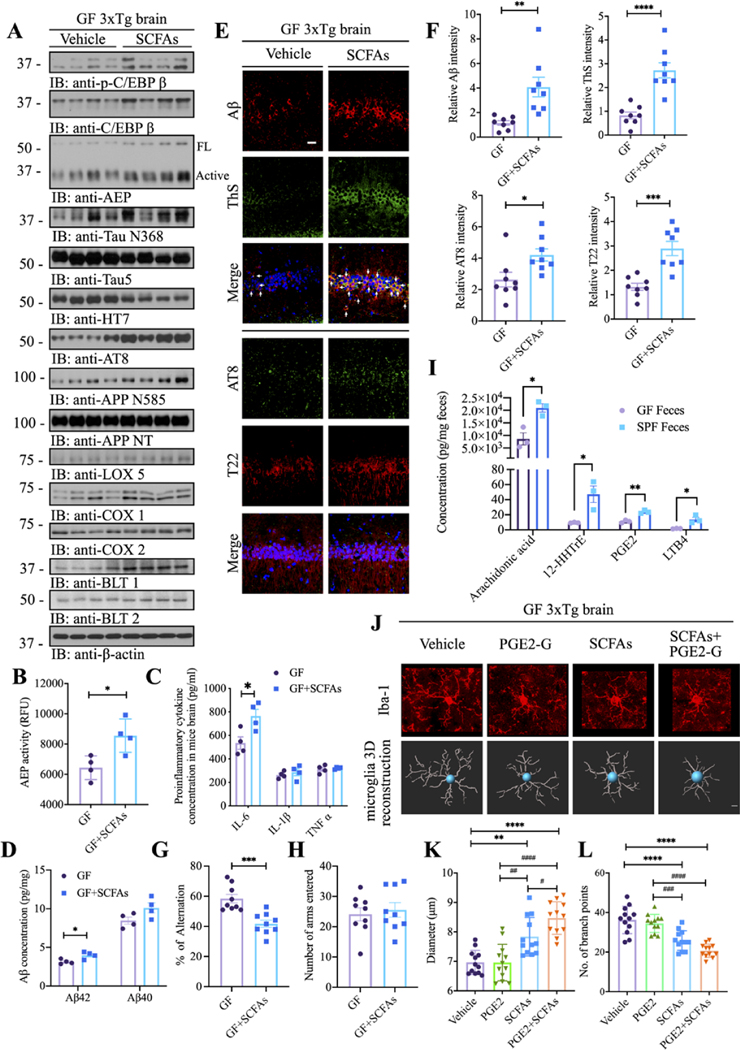
AD patients’ gut microbiota possess lower proportions of key butyrate-producing species, such as members of Butyrivibrio and Eubacterium genera. AD elders also display diminished butyrate enzyme encoding genes than the healthy elders without dementia. Lower proportions of butyrate producing species would lead to a proinflammatory state.12 Indeed, short chain fatty acids (SCFAs) were decreased in the faeces and the serum from AD patients’ faecal transplanted ex-GF mice (see below online supplemental figure 11). To investigate whether SCFAs would partially mimic the gut microbiomes in colonised SPF mice to trigger C/EBPβ/AEP pathway activation, we fed GF 3×Tg mice with vehicle control or SCFAs with a mixture of acetate, propionate and butyrate (while the animals remained microbiologically sterile) consecutively for 3 months. Immunoblotting showed that p-C/EBPβ and its total protein levels were escalated in GF mice fed SCFAs; consequently, the downstream AEP and its active form were increased, which triggered Tau N368 and APP N585 augmentation. Subsequently, p-Tau AT8 activities were accordingly elevated. As a result, the downstream targets of C/EBPβ, including Lox-5, Cox1, Cox2, BLT1 and BLT2 levels were augmented by SCFA feeding as compared with vehicle (figure 4A). Quantification of enzymatic assay confirmed that AEP protease activities were significantly enhanced in SCFA-fed GF mice vs vehicle (figure 4B). In alignment with C/EBPβ activation, the downstream responsive inflammatory cytokine IL-6 was increased (figure 4C). Aβ ELISA quantification revealed that Aβ42 but not Aβ40 concentrations in the brains were prominently increased by SCFA treatment (figure 4D). These findings were further corroborated by IF costaining. In the hippocampus, ThS-positive Aβ aggregates and p-Tau AT8 and fibrillar Tau T22 signals were intensified by SCFAs (figure 4E,F). In addition, elevated AEP, associated with AEP-truncated Tau N368 and APP C586 fragments were also enriched on SCFAs treatment (online supplemental figure 6A–F). Consistent with the engendered AD pathologies in GF 3×Tg mice by SCFAs, cognitive behavioural tests showed that percentage of alterations was significantly suppressed in SCFA fed GF mice, though the number of arms entered remained unchanged (figure 4G,H). Microglia, the resident macrophages of the brain, protect the CNS from injury and infection. Microglia also contribute to the brain development throughout embryonic and adult life by regulating aspects of synaptic transmission, synapse formation and pruning and embryonic brain wiring.39 Within the affected hippocampus and cortex regions, microglia in SCFA-administered GF mice exhibited morphology indicative of increased activation and maturation compared with vehicle-treated GF mice, and similar to microglia cells from GF and SPF mice, respectively (figure 4J, first and third panels).
Figure 4.
SCFAs trigger C/EBPβ/AEP activation and cognitive deficits in GF 3xTg mice and inflammation, exacerbated by PGE2-G. (A) Immunoblot showing p-C/EBPβ, C/EBPβ, AEP, APP and tau expression and processing, as well as arachidonic acid metabolism in the brains of GF mice with or without SCFAs administration. (B) AEP activity assay in the brain lysates from GF 3xTg mice with or without SCFAs administration. SCFAs treatment elevates AEP activity in the brains of GF 3xTg mice. Data represent the mean±SEM; representative data of four samples; *p<0.05 compared with control, unpaired t-tests. (C) Proinflammatory cytokine IL1-β, IL-6 and TNFα concentrations in the brain lysates from GF 3xTg mice and SPF 3xTg mice, respectively. Data represent the mean±SEM; representative data of four samples; *p<0.05 compared with control, multiple unpaired T tests. (D) Aβ40 and Aβ42 concentrations in the cortex of GF 3xTg mice with or without SCFAs administration were measured using human aβ40 and Aβ42 ELISA kit. The concentrations of Aβ42 not aβ40 were significantly increased in the cortex regions of the brains from SCFAs-treated GF 3xTg mice compared with vehicle-treated GF 3xTg mice. (N=4 in each group, data are shown as mean±SEM *p<0.05 compared with control, multiple unpaired T tests). (E) Immunofluorescent staining of Aβ (red) and ThS (green), AT8 (green) and T22 (red) in the hippocampus CA1 region of brains from GF 3xTg mice with or without SCFAs administration. Scale bar: 20 μm (F) quantitative analysis of Aβ positive cells, ThS positive cells, AT8 positive cells and T22 positive cells, respectively. The density of Aβ, ThS, AT8 and T22 positive cells was significantly increased in SCFAs-treated GF 3xTg mice brains. (n=8 in each group, data are shown as mean±SEM. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001 compared with control, unpaired T tests.). (G, H) Y-maze behavioural tests. spontaneous alternation (%) (G), number of arms entered (H); (N=9 in each group, data are shown as mean±SEM. ***P<0.001 compared with control, unpaired T tests). (I) Arachidonic acids and its metabolites concentrations in the faeces of GF mice and SPF mice. (N=3 in each group, data are shown as mean±SEM, *p<0.05, **p<0.01 compared with control, multiple unpaired T tests). (J) Representative images of immunofluorescent staining of Iba-1 (red) in the cortex (upper panel) and 3D reconstruction of Iba-1-stained microglia (lower panel) residing in the cortex of GF 3xTg mice, GF 3xTg mice treated with either PGE2-G or SCFAs, GF 3xTg mice treated with both SCFAs and PGE2-G. (K, L) Quantitative analysis of diameter, number of branch points of microglia residing in the cortex. data represent the mean±SEM; representative data of twelve samples; **p<0.01, ***p<0.001, ****p<0.0001 compared with control, multiple unpaired T tests; #P<0.05, ##P < 0.01, ###P < 0.001, #### P < 0.0001 compared with control, multiple unpaired t tests. Aβ, amyloid-β; AEP, asparagine endopeptidase; GF, germ-free; PGE2-G, prostaglandin E2–1-glyceryl ester; SCFA, short chain fatty acids.
Metabolomic studies on the brain tissue samples provide direct insights into the molecular basis of AD pathogenesis. Accordingly, we performed metabolomics analysis with the brain samples from GF and SPF 3×Tg mice. Metabolomics revealed AA-associated metabolites in the brain were diminished in GF 3×Tg mice versus SPF mice (online supplemental figure 7). As expected, PUFA metabolites were significantly reduced in the faeces from GF mice as compared with SPF (figure 4I). Consequently, we chose PGE2-G, a glyceryl-conjugated metabolite from PGE2, which was identified from the metabolomics analysis. PGE2-G treatment barely elicited immature microglia activation in GF mice vs vehicle. Nevertheless, SCFA and PGE2-G cotreatment additively stimulated microglia morphology maturation and activation in GF mice (figure 4J–L), indicating that PGE2 stimulates mature but not immature microglia activation. Together, these findings support that SCFAs trigger C/EBPβ/AEP activation and cognitive deficits in GF 3×Tg mice and inflammation, exacerbated by AA-metabolite PGE2-G.
Bacteroides strains implicated in PUFA metabolism are escalated in AD microbial recolonised GF mice
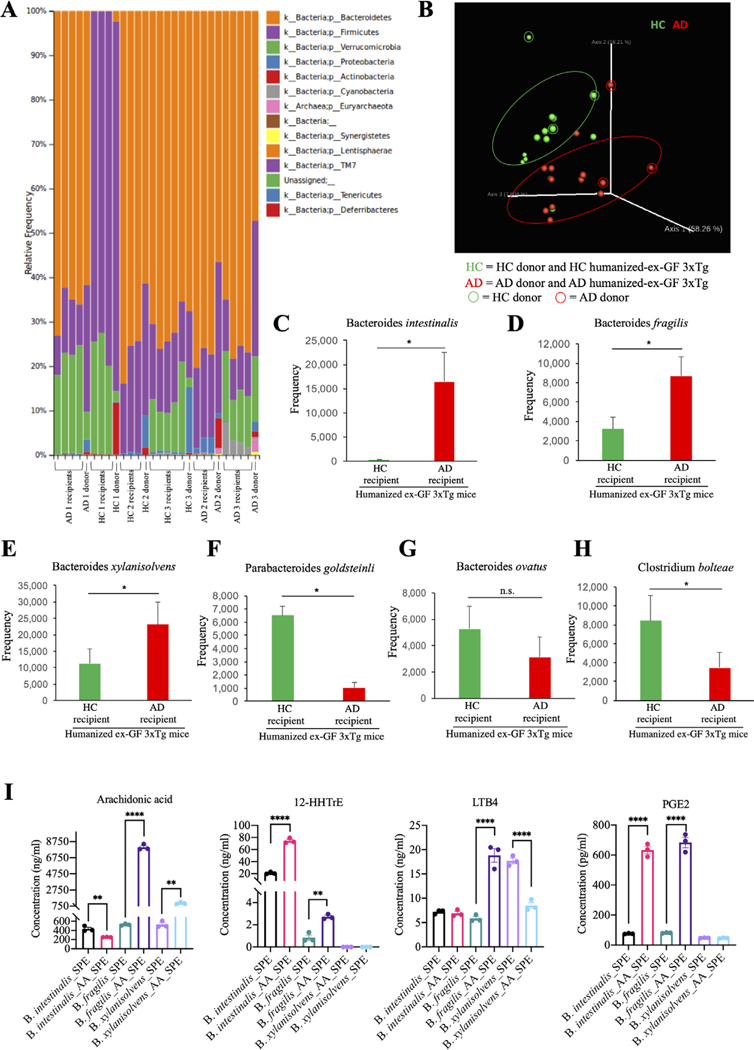
Since patients with AD display altered microbiomes,10–12 we sought to determine whether human-derived gut microbiomes affect disease outcomes when transferred into GF mice. We inoculated three human AD and three HC donors’ faecal samples into GF mice via oral gavage. The faecal pellets were collected from humanised mice, bacterial DNA extracted, and 16S rRNA sequencing was conducted. The microbiome analysis revealed an establishment of the human gut microbiota by faecal microbiota transplant (FMT) in ex-GF mice. At the phyla level, the taxonomic analysis demonstrated a strong resemblance of the microbial community of FMT-recipient mice with the microbial phyla of the human donor inoculum (figure 5A). As shown in several previous studies, the recipient mice successfully harboured 100% phyla-level and 86% of the genus-level taxa, which was detected in the donor inoculum. Furthermore, the beta diversity analysis PCoA showed that the microbiota of the FMT-recipient mice was clustered together with their human donor counterparts (figure 5B). Importantly, our study confirmed that there were no significant alterations in the alpha diversity between the donor and FMT recipient mice. Hence, our data suggest successful engraftment of human gut microbiota in the GF mice. In addition, our findings demonstrated a remarkable escalation in the mean relative abundances of Bacteroides intestinalis (figure 5C), Bacteroides fragilis (figure 5D) and Bacteroides xylanivsolvens (figure 5E), and a significant decrease in the mean relative abundances of Parabacteroides goldsteiniii (figure 5F), Bacteroides ovatus (figure 5G) and Clostridium bolteae (figure 5H). Strikingly, quantitative analysis of the concentrations of AA and its metabolites in culture medium from the three strains of bacteria (B. intestinalis, B. fragilis and B. xylanivsolvens) increased in AD humanised ex-GF mice indicated that these bacteria are involved in AA metabolism and can generate AA metabolites (figure 5I).
Figure 5.
Microbiome analysis in humanised ex-germ-free 3xTg mouse stool, revealing bacteroides elevation in the faeces from human AD faecal inoculated germ-free mice. (A) relative abundance of bacterial phyla determined by high throughput sequencing analysis. (B) principal coordinate plot (PcoA) of microbial community structure. (C–H) Mean frequency of bacterial species. Data represent the means±SEM; *p<0.05 compared with control, one-way ANOVA. (I) Concentrations of arachidonic acid (AA) and its metabolites in culture medium from in vitro culture of Bacteroides intestinalis, Bacteroides fragilis and Bacteroides xylanisolvens. Data represent the means±SEM; representative data of three samples; **p<0.01, ****p<0.0001 compared with control, one-way ANOVA. AD, Alzheimer’s disease; ANOVA, analysis of variance.
AD patient gut microbiota enhance AD pathology and promote cognitive deficits
To assess whether human AD faecal samples could accelerate AD pathologies in GF mice, we performed IF costaining and found that ThS-positive Aβ aggregates were significantly elevated in AD humanised ex-GF 3×Tg mouse brain as compared with HC. Moreover, AT8/T22 costaining in the hippocampus also exhibited the similar pattern (figure 6A,B), supporting that AD patient microbiomes facilitate AD pathologies including senile plaques and NFT in GF 3×Tg mice. Quantification showed that Aβ42 but not Aβ40 concentrations were substantially increased in AD faecal inoculated ex-GF mice (figure 6C). IF costaining revealed that C/EBPβ and AEP were robustly elevated in AD faecal inoculated ex-GF mice as compared with HC; consequently, Tau N368, APP C586 and Aβ were all greatly augmented, coupled with noticeable microglial activation, validated by prominent Iba-1 and CD86 costaining signals (online supplemental figure 8A–E,H). Consistent with upregulation of AA metabolic enzymes, LKB4 and 12-HHT receptors, BLT1 and BLT2, two downstream transcriptional targets of C/EBPβ,40 were also pronouncedly enhanced in human AD microbial transplanted ex-GF mice (online supplemental figure 8F–H). In agreement with the prominent AD pathological features, immunoblotting revealed that synaptic proteins including PSD95, GluR2 and Spinophilin were decreased in AD faecal transplanted ex-GF mice versus HC (online supplemental figure 9A). Compatible with these findings, BDNF levels were significantly lowered in AD faecal humanised mice (online supplemental figure 9B). Consistently, the dendritic spines in the hippocampal neurons were greatly reduced (online supplemental figure 9C,D). Consequently, AD faecal humanised mice exhibited cognitive deficits in the Y-maze, as compared with HC mice, though the sum number of the arms entered was not statistically changed. However, one pair of the ex-GF mice inoculated with AD versus HC faecal samples exhibited significantly reduced arm entry (figure 6D–G). As expected, Iba-1 staining, and 3D microglial reconstruction analysis demonstrated the microglia cells in AD subject-inoculated GF mice were highly activated and morphologically mature (figure 6H–K). Therefore, these data support that differences in faecal microbial communities between patients with AD and HCs might be maintained after inoculated into GF mice.
Figure 6.
AD faecal humanised ex-germ-free (GF) mice exhibit augmented AD pathologies and cognitive deficits. (A) immunofluorescent staining of Aβ (red) and ThS (green) in the frontal cortex region of brains, AT8 (green) and T22 (red) in the hippocampus CA1 region of the brains from HC humanised ex-GF 3xTg mice and AD humanised ex-GF 3xTg mice. HC, GF mice colonised with faecal microbiomes from HC; AD, GF mice colonised with faecal microbiomes from AD patients. Scale bar: 20 μm (B) quantitative analysis of Aβ positive cells, ThS positive cells, AT8 positive cells and T22 positive cells, respectively. The density of Aβ, ThS, AT8 and T22 positive cells was significantly increased in AD humanised ex-GF 3xTg mice brain. (N=5 in each group, data are shown as mean±SEM **p<0.01, ***p<0.001, ****p<0.0001 compared with control, unpaired T tests). (C) aβ40 and Aβ42 concentrations in the cortex of HC humanised ex-GF 3xTg mice and AD humanised ex-GF 3xTg mice were measured using human aβ40 and Aβ42 ELISA kit. The concentrations of Aβ42 not aβ40 were significantly increased in AD humanised ex-GF 3xTg mice cortex compared with HC humanised ex-GF 3xTg mice cortex. (N=5 in each group, data are shown as mean±SEM ***p<0.001 compared with control, multiple unpaired T tests). (D–G) Y-maze behavioural tests. (D–E) compilation of all independent cohorts in each Y-maze test: spontaneous alternation (%), number of arms entered, grouped by healthy status of faecal donor (n=23–24 in each group, data are shown as mean±SEM. *P<0.05 compared with control, unpaired t-tests); (F–G) spontaneous alternation (%), number of arms entered of mice humanised with microbiota from either AD patients or matched HCs. (N=7–8 in each group, data are shown as mean±SEM, *p<0.05 compared with control, unpaired t-tests). (H) Representative images of immunofluorescent staining of Iba-1 (red) in the cortex (upper panel) and 3D reconstruction of Iba-1-stained microglia (lower panel) residing in the cortex of HC humanised ex-GF 3xTg mice and AD humanised ex-GF 3xTg mice. (I–K) quantitative analysis of diameter, number of branch points, and total branch length of microglia residing in the cortex. data represent the mean±SEM; representative data of 12 samples; *p<0.05, **p<0.01 compared with control, unpaired t-tests. Aβ, amyloid-β; AD, Alzheimer’s disease; HC, healthy control.
AD fecal humanised ex-GF mice demonstrate elevated C/EBPβ/AEP activation, associated with elevated inflammation
To characterise in-depth the pathological events in the humanised ex-GF 3×Tg mice, we conducted immunoblotting and found that p-C/EBPβ and its total protein levels were pronouncedly elevated by human AD microbiomes transplantation as compared with HC. The downstream AEP and its mature form also oscillated with the upstream transcription factor activities. Consequently, Tau N368 and APP N585 and p-Tau AT8 signals were prominently elevated. Again, the downstream responsive genes including Lox-5, Cox1, Cox2, BLT1 and BLT2 levels all echoed the upstream C/EBPβ signals (figure 7A,B). As expected, AEP enzymatic activities were appropriately enhanced (figure 7C). Quantification of inflammatory cytokines showed that IL-6 and IL-1β were significantly enhanced by human AD faecal transplantation (figure 7D). qRT-PCR demonstrated that ALOX-5 mRNA concentrations not other AA-metabolism-associated genes were highly increased (figure 7E), indicating that ALOX5 promoter might be more sensitive and responsive to the upstream transcription factor C/EBPβ activity. These findings correlate with the observation that AA metabolites were augmented in AD microbial re-GF mice (online supplemental figure 11).
Figure 7.
AD faecal humanised ex-GF mice demonstrate elevated C/EBPβ/AEP activation, associated with elevated inflammation. (A) Immunoblot showing p-C/EBP β, C/EBP β, AEP, APP and tau expression and processing, as well as arachidonic acid metabolism in mouse brains of HC humanised ex-GF 3xTg mice and AD humanised ex-GF 3xTg mice. Four samples in each group are from two separated donors (2 samples per donor). (B) Quantitative analysis of immunoblot. The bands of C/EBPβ, LOX5, COX1, COX2, BLT1 and BLT2 were measured with Image J and normalised with β-actin. (N=4 in each group, Data are shown as mean±SEM, *p<0.05, **p<0.01 compared with control, unpaired t tests). (C) AEP activity assay in the brain lysates from HC humanised ex-GF 3xTg mice and AD humanised ex-GF 3xTg mice. AD humanised ex-GF 3xTg mice showed escalation in AEP activity compared with that in HC humanised ex-GF 3xTg mice. Data represent the mean±SEM; representative data of five samples; *p<0.05 compared with control, unpaired t-tests. (D) Proinflammatory cytokine IL1-β, IL-6 and TNFα concentrations in the brain lysates from HC humanised ex-GF 3xTg mice and AD humanised ex-GF 3xTg mice, respectively. Data represent the mean±SEM; representative data of five samples; *p<0.05 compared with control, multiple unpaired t-tests. (E) Relative mRNA levels of genes involving arachidonic acid metabolism in the brain from HC humanised ex-GF 3xTg mice and AD humanised ex-GF 3xTg mice. Data represent the mean±SEM; representative data of three samples, *p<0.05 compared with control, two-way ANOVA. AD, Alzheimer’s disease; AEP, asparagine endopeptidase; ANOVA, analysis of variance; COX, cyclooxygenases; GF, germ-free; HC, healthy control.
The PCA analysis suggested that there were some metabolic differences within the HC and AD groups humanised ex-GF as well as within the GF and SPF groups (online supplemental figure 10A–C). Remarkably, the metabolomic profiles for the faeces and serum samples were significantly different in respect to AD humanised ex-GF compared with HC humanised ex-GF mice. While the metabolomic profiles for the brains appeared significantly different with respect to SPF compared with GF mice, the metabolic differences seem minimal regarding AD humanised ex-GF compared with HC humanised brains. Within the faeces dataset, several aromatic metabolites were significantly higher or trending higher (not significant) in the AD compared with the HC humanised ex-GF group, while secondary bile acids (eg, taurolithocholate 3-sulfate), vitamins (eg, thiamin monophosphate), and benzoate metabolites (eg, benzoate) were significantly altered or trending altered (not significant) (online supplemental figure 10D). In the serum, several aromatic amino acid metabolites (eg, phenylpyruvate, dopamine 3-O-sulfate and 3-indoxyl sulfate), and secondary bile acids (eg, taurohyodeoxycholic acid), vitamins (eg, pyridoxal) and benzoate (eg, 4-ethylphenylsulfate) metabolites were significantly higher in the AD compared with the HC humanised ex-GF group (online supplemental figure 10E). Further, within the brain, dopamine was trending lower, while the benzoate metabolites hippurate and catechol sulfate were significantly higher or trending higher, respectively, in the AD compared with the HC humanised ex-GF group (online supplemental figure 10F). Several pentose metabolites were significantly higher in the AD humanised ex-GF group in faeces (online supplemental figure 10G). In the serum, glycolytic metabolite pyruvate was significantly higher, while pentose, TCA cycle and the NAD metabolites displayed similar results as the faeces when comparing the AD with the HC humanised ex-GF group (online supplemental figure 10H). The TCA energy metabolites aconitate and malate were significantly higher in the AD humanised ex-GF group in the brain (online supplemental figure 10I). For lipid metabolism, phosphatidylethanolamine (PE) metabolites and 1-stearoyl-2-linoleoyl-GPE were higher in the faeces of the AD humanised ex-GF group (online supplemental figure 11A). Several LCFAs (long-chain fatty acids, eg, myristoleate and docosapentaenoate) as well as several carnitine-conjugated fatty acids were significantly lower within the AD humanised ex-GF group in the serum (online supplemental figure 11B). Notably, SCFAs, especially butyrate, were reduced in the faeces and serum (online supplemental figure 11A,B). Sphingomyelin related metabolites were significantly higher in the AD humanised ex-GF group compared with the HC in the brain (online supplemental figure 11C). AA-associated metabolites in the serum and brain, including Thromboxane B2, PGF2α and 12-HHTrE etc, were increased but not significantly. These data are suggestive of significant alterations in lipid metabolism in the AD humanised ex-GF group compared with the HC humanised ex-GF group. Arginine contributes to multiple aspects of cellular metabolism, inflammatory signalling and energy metabolism. Arginine metabolism is altered in AD patients.41 Within the faeces, serum and brain dataset, several arginine metabolites were significantly higher in the AD humanised ex-GF group compared with the HC group (online supplemental figure 11D–F).
Immunoblotting also revealed that Lox5, Cox1, Cox2, BLT1 and BLT2 levels were much more abundant in human AD patient brains compared with age-matched control brains (online supplemental figure 12A,B), consistent with elevated AA-metabolites in the serum and the brains from AD humanised ex-GF mice. IF costaining further demonstrated that C/EBPβ and Lox5 were escalated in Iba-1-positive microglia cells and NeuN-positive neurons in human AD brains vs control brains. Additionally, marked C/EBPβ augmentation in human AD brains was accompanied with enhanced Lox5 and AEP signals, two downstream transcriptional targets (online supplemental figure 12C,D). Hence, human AD microbiomes augment AD pathologies and AA metabolic enzymes in transplanted ex-GF mice, associated with inflammatory microglia activation.
DISCUSSION
Gut microbiota plays a major role in peripheral as well as central immune activation and inflammation in various neurological disorders. In the current study, we provide evidence showing that the gut microbiome is required for AD pathology and cognitive deficits in an AD mouse model. Under the GF conditions, 3×Tg mice display reduced microglia activation, senile plaques and NFT, and spatial memory defects as compared with SPF animals. RNAseq reveals that insulin signalling is aberrantly altered and proinflammatory pathways are elevated in the SPF vs GF 3xTG mouse brains. Further, metabolomic analysis from humanised GF mouse brains, serum and faeces identifies proinflammatory signalling escalation and numerous AA-related metabolites, associated with SCFAs reduction. Treatment with PGE2-G, a metabolite from AA by bacterial strain Bacteroides, restores AD pathologies and microglial activation in GF mice in the presence of SCFA. This finding provides a potential molecular mechanism driven by the gut–brain axis. Impaired spatial memory in humanised mice transplanted with AD microbiota compared with HC indicates that Aβ and Tau overexpression and dysbiosis interact and modulate the disease progression. Conceivably, these preclinical findings might be applied to humans and the gene–microbiome interactions may provide insight into AD pathogenesis.
C/EBPβ is a key player in the regulation of gene expression in neuroinflammation.42 Accordingly, we found that it is activated in the microglia cells in the brains from SPF or AD patient faecal transplanted mice and escalates mRNA transcription of the major enzymes implicating in AA metabolism (figures 2 and 7, (online supplemental figure 4 and online supplemental figure 8). These findings are consistent with previous reports that the expression of COXs and LOX5 and PTGES are mediated by C/EBPβ.42–45 Ptges3 encodes cytosolic prostaglandin E synthase (cPGES), and Pgtes2 encodes microsomal PGES-2 (mPGES-2), both cPGES and mPGES-2 are important PGES to convert PGH2 to PGE2. cPGES preferentially coordinates with COX-1 to convert PGH2 to PGE2, while mPGES-2 could couple with both COX-1 and COX-2 to produce PGE2 in response to inflammation.46 Less inflammation and PGE2 in GF mice are consistent with the reduced Hpgd and Pgtes3 mRNA levels. While mPGES-2 is constitutively expressed in tissue and is not increased appreciably during tissue inflammation or damage.47 Presumably, because PGE2 level is lower in GF mice, Pgtes2 may increase as a feedback compensation (figure 3D). Alox12 gene-encoded enzyme and its reaction products regulate platelet function. Elevated expression of this gene has been observed in pancreatic islets derived from human diabetes patients,48 but its role in the brain is unclear. Thus, it remains unknown whether it is mediated by C/EBPβ or not. The elevated enzymes lead to upregulation of various AA metabolites including PGE2, Thromboxane B2, LKB4 and 12-HHT etc., which feedback to stimulate microglia activation, further escalating neuroinflammation. Moreover, C/EBPβ regulates numerous proinflammatory genes in glial activation.49 In alignment with these findings, PGE2-G and SCFA treatment elicits prominent microglia activation and AD pathologies in GF 3xTg mice (figure 4). Hence, there is a vicious cycle between C/EBPβ and enzymes mediating proinflammatory AA metabolism to amplify neuroinflammation. In addition to glia cells, C/EBPβ also acts as a pivotal transcription factor in neurons regulating the expression of major AD effectors including APP, MAPT and AEP etc.31 Recently, we reported that C/EBPβ/AEP signalling is age-dependently escalated in neurons and augmented APP and Tau proteins are subsequently cleaved by active AEP, triggering AD pathogenesis.31 35 50 Interestingly, C/EBPβ deficiency provides neuroprotection following ischaemic or excitotoxic injuries,51 two models where COX-2 inhibition is beneficial.52 53 Presumably, C/EBPβ/AEP signalling activation in the gut of 5×FAD15 or in the brains of SPF mice and GF mice humanised by AD faecal samples is partially mediated by the proinflammatory AA metabolite due to gut dysbiosis in AD subjects (figures 2 and 7).
RNAseq analysis of SPF and GF mouse brains reveals several crucial signalling pathways are implicated in AD pathologies. For instance, the insulin/IGF-1 pathway and its downstream effectors PI3-kinase/Akt (PKB) and RAS/RAF/MAP Kinase etc. are pronouncedly altered in SFP mice as compared with GF mice (figure 3). Interestingly, AD is also called type 3 diabetes (T3D). Patients with AD show low levels of insulin, IR, and overall insulin signalling impairments in their brains. Also, there is an increased chance of developing mild cognitive impairment (MCI), dementia, or AD in T2D mellitus (T2DM) individuals. Furthermore, patients with AD show progressive brain insulin resistance and insulin deficiency.37 38 Animal models of T2DM or obesity display the cognitive impairment. Treating with insulin sensitiser agents or intranasal insulin improves cognitive performance in animal models and humans with AD and MCI.54 We realise that it is not accurate to draw parallel between AD versus HC and GF versus SPF mice. However, the insulin signalling changes between 3×Tg mice and wild-type mice have been well studied from previous research. Mounting evidence shows impairment of glucose tolerance in the 3×Tg mice, including female 3×Tg mice displaying a significant deterioration in glucose tolerance, glucose intolerance and cortical amyloid pathway worsened with age.55 Chronic treatment of insulin sensitisers improved spatial learning, and attenuated tau hyperphosphorylation and neuroinflammation.56 Previous studies demonstrate that insulin signalling and C/EBPβ mutually regulate each other. For example, insulin triggers numerous stress-responsive gene expression via inducing C/EBPβ.57 Notably, C/EBPβ implicates in mediating IGF-1 and human IR expression via binding their promoters.58 59 Further, C/EBPβ is indispensable for IGF-II-mediated memory consolidation and enhancement,60 and PGE2 upregulates IGF-1 expression via activating C/EBPβ.61 62 Hence, these observations support that gut microbiota might trigger C/EBPβ/AEP pathway activation in the brain via AA metabolites, leading to abnormal insulin/IGF-1 signalling in AD mouse brains.
Metabolomics and HPLC analysis indicate that the AA pathway’s proinflammatory metabolites are strongly enhanced in the brain and faecal samples from the SPF versus GF mice and AD vs HC faecal transplanted mice (figures 4I and 5I, (online supplemental figures 7 and 11). These observations are consistent with previous reports that AA, PGE2 and LKB4 etc. are elevated in the faecal samples from patients with AD as compared with HCs, and AA metabolism is elevated in an AD mouse model22 and AD patient brains.21 63 Remarkably, we also found that bacteria strain Bacteroides that are implicated in AA metabolism is highly enhanced in re-colonised GF mice with human AD faecal samples (figure 5C–E). Conceivably, the intestinal bacteria metabolise AA into PGE2 (figure 5I), which may enter the circulation system due to the gut leakage, triggered by gut dysbiosis. The metabolites may penetrate into the brain due to BBB impairments in 3×Tg mice, stimulating microglia activation and provoking neuroinflammation. Previous report also shows that gut microbiota damages the integrity of BBB, leading to the leakage of BBB and endotoxins infiltration.64
Several metabolomic studies have examined the relationship between metabolism and AD pathology. Studies on serum, plasma, and cerebrospinal fluid (CSF) have identified numerous metabolic pathways involving bile acids, sphingolipids, antioxidants, phospholipids, and amino acids that appear to be associated with disease.65 A number of studies with brain tissue samples from both transgenic animal models and humans show alterations in neurotransmitters, amino acids and antioxidants to be strongly associated with AD66 and increased brain polyamine metabolism in AD brains.67 Interestingly, dopamine was significantly lower in the AD humanised ex-GF mouse brains (online supplemental figures 7C and 10F), fitting with previous report that dopamine is lower in AD patients.68 Alterations in aromatic amino acid metabolites in addition to secondary bile acids, vitamins and benzoate metabolites suggest an altered state of microbial metabolism and/or microbial populations in the AD humanised ex-GF group compared with the HC humanised ex-GF group, supporting that administration of human AD patient faeces altered the microbiome in the AD humanised ex-GF group. Pentose sugars are metabolised within the gut by microbial action providing carbon and energy69 and alterations of pentose metabolites indicates an altered state of microbial metabolism and/or microbial populations within the AD humanised ex-GF group compared with the HC humanised ex-GF group. In addition, differences in glycolytic and TCA cycle metabolites are suggestive of an alteration in glucose utilisation, energy homeostasis, and excretion. Higher levels of brain phospholipids coupled with decreased serum LCFAs and carnitine-conjugated lipids, and increased faeces PE lipids could perhaps, suggest altered lipid metabolism in AD humanised GF group (online supplemental figures 10 and 11). Increased levels of ornithine could be indicative of decreased nitric oxide production, which is supported by the observed higher levels of polyamine metabolites in serum (N-acetylputrescine) and in faeces (N-carbamoylputrescine) in the AD humanised ex-GF group compared with the HC humanised ex-GF group. Notably, there are indications of differences in gut microbial metabolism as evident from the changes observed in the following classes of metabolites: aromatic amino acid metabolites, secondary bile acids, vitamins and benzoates. Additionally, there were differences in carbohydrate and energy metabolites, lipid metabolites, arginine metabolites and oxidative stress metabolites. Interestingly, indoxyl sulfate, a metabolite from dietary tryptophan, is selectively elevated in the serum and the brains of AD-humanised ex-GF mice vs HC and the brains from SPF mice versus HG mice (online supplemental figures 7 and 10). It disrupts BBB integrity, increases oxidative stress and elevates neuroinflammation by escalating inducible nitric oxide and COX-2, TNFα and IL-6 in glial cells.70 Conceivably, it may facilitate the inflammatory metabolites like PGE2-G penetrates the brain to trigger microglia activation.
Indisputably, the mechanisms by which gut bacteria promote Aβ or Tau-mediated pathologies might be complex; in the current report, we have identified one potential pathway requiring microbiota-dependent effects on microglia. Microglial maturation and function are impaired both in the absence of the microbiome in GF or ABX (antibiotics)-treated mice.71 72 For example, microglial cells in GF and ABX-treated mice display significantly altered inflammatory gene expression profiles that influence their surveillance function that is accompanied by altered cellular morphologies. Long-term ABX treatment resulted in reductions of Aβ deposition and microglial morphological alterations in male but not female APP/PS1 mice.73 Here, we show that microglia cells display different morphologies in GF mice compared with SPF mice (figure 1F–I), indicating that microglia maturation is impaired in GF mice, which is consistent with previous findings.71 Microbial re-colonisation of GF mice with AD patient faecal samples activates microglia as compared with healthy donors (figure 6H–K). SCFAs, microbiota-derived bacterial fermentation products, regulate microglia homeostasis.71 AD patients gut microbiota possess lower proportions of key butyrate-producing species, such as members of Butyrivibrio and Eubacterium genera. AD elders also display diminished butyrate enzyme encoding genes than the healthy elders without dementia.12 Hence, our observation fits with these findings. SCFAs treatments escalate the microglia maturation in GF mice as compared with untreated mice, and addition of PGE2-G metabolite further increases microglia activation (figure 4J–L). Butyrate is an essential metabolite that is the preferred energy source for the colonic epithelial cells, and it contributes to the gut barrier maintenance and has both immunomodulatory and anti-inflammatory properties.74 Conceivably, lack of antiinflammatory butyrate-producing bacteria in AD gut microbiota, combined with elevated inflammatory PGE2 metabolite in the brain, additively augment microglial activation, resulting in chronic neuroinflammation. This subsequently activates C/EBPβ/AEP pathway, which elevates APP and tau expression levels and enhances AEP δ-secretase activity, leading to AD pathogenesis.
Experimental procedures
Key resources table 1.
Table 1.
Key resources
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Anti-δ-secretase (AEP) (11B7) | Dr. Colin Watts (University of Dundee) | N/A |
| Anti-TauN368 | Home-made | N/A |
| Anti-APPC586 | Home-made | N/A |
| Anti-APPN585 | Home-made | N/A |
| Anti-C/EBPβ(H-7) HRP | Santa Cruze | Cat# sc-7962 HRP |
| Anti-phoshpo Thr235 C/EBPβ | Cell Signalling Technology | Cat# 3084s |
| Anti-beta Amyloid | Sigma-Aldrich | Cat#A8354 |
| Anti-Tau (T22) oligomeric | Millipore Sigma | Cat# ABN454 |
| Anti-phospho-Tau (Ser202, Thr205) (AT8) | Thermo-fisher | Cat# MN1020 |
| Anti-phospho-Tau (Thr212, Ser214) (AT100) | Thermo Fisher | Cat# MN1060 |
| Anti-Iba1 | VWR | Cat# 1 00 369–764 |
| Anti-NeuN | Invitrogen | Cat# PA5-78639 |
| Anti-δ-secretase (AEP) (D6S4H) | Cell Signalling Technology | Cat# 93 627 |
| Anti-CD86 (E2G8P) | Cell Signalling Technology | Cat# 91 882S |
| Anti-Tau (TAU-5) | Invitrogen | Cat# AHB0042 |
| Anti-Tau (HT7) | Invitrogen | Cat# MN1000 |
| Anti-APP A4 (22 C11) | Millipore Sigma | Cat# MAB348 |
| Anti-5 Lipoxygenase (LOX5) | Abcam | Cat# ab39347 |
| Anti-Cyclooxygenase 1 (COX1) | Abcam | Cat# ab109025 |
| Anti-COX2 | Abcam | Cat# ab15191 |
| Anti-LTB4-R1/BLTR | Thermo Fisher | Cat# BS-2654R |
| Anti-LTB4-R2 | Thermo Fisher | Cat# BS-2655R |
| Anti-β actin (AC-15) | Invitrogen | Cat# AM4302 |
| Anti-mouse IgG-HRP | Cell Signalling Technology | Cat# 7076S |
| Anti-rabbit IgG-HRP | Cell Signalling Technology | Cat# 7074S |
| Anti-mouse IgG-Alexafluor 488 | Invitrogen | Cat# A11001 |
| Anti-mouse IgG-Alexafluor 594 | Invitrogen | Cat# A11005 |
| Anti-rabbit IgG-Alexafluor 488 | Invitrogen | Cat# A11034 |
| Anti-rabbit IgG-Alexafluor 594 | Invitrogen | Cat# A11037 |
| Anti-rabbit IgG-Cy5 | Invitrogen | Cat# A10523 |
| Chemical and Peptides | ||
| Thioflavin S | Millipore Sigma | Cat# T1892-25G |
| Prostaglandin E2-1-glyceryl ester | Cayman chemical | Item No. 10 140 |
| Arachidonic acid | Cayman chemical | Item No. 90 010 |
| PGE2 | Cayman chemical | Item No. 14 010 |
| LTB4 | Cayman Chemical | Item No. 20 110 |
| 12-HTTrE | Cayman chemical | Item No. 34 590 |
| Sodium Acetate | Sigma-Aldrich | Cat# S2889-1KG |
| Sodium Propionate | Sigma-Aldrich | Cat# P1880-1KG |
| Sodium Butyrate | Sigma-Aldrich | Cat# 3 03 410-500G |
| Vancomycin Hydrochloride | VWR | Cat# 97 062-554 |
| Gentamicin sulfate | Sigma-Aldrich | Cat# G1914-5G |
| Ampicillin | Sigma-Aldrich | Cat# A159325 |
| Erythromycin | Sigma-Aldrich | Cat# PHR1039-1G |
| Neomycin | Fisher Scientific | Cat# 21 810 031 |
| Z-Ala-Ala-Asn-AMC (AEP substrate) | BACHEM | Cat# I-1865 |
| Critical Commercial Assay | ||
| Amyloid beta 42 Human ELISA Kit | Invitrogen | Cat# KHB3441 |
| Amyloid beta 40 Human ELISA Kit | Invitrogen | Cat# KHB3481 |
| Mouse IL-6 ELISA Kit | Abcam | Cat# ab100712 |
| TNF alpha Mouse Uncoated ELISA Kit | Invitrogen | Cat# 88-7324-88 |
| IL-1 beta Mouse Uncoated ELISA Kit | Invitrogen | Cat# 88-7013-22 |
| PGE2 ELISA Kit | Abcam | Cat# ab133021 |
| LTB4 ELISA Kit | Invitrogen | Cat# EHLTB4 |
| Deposited Data | ||
| 16 s RNA sequences | PPMS for EIGC https://fex.cores.emory.edu/ | KYE22363-63838 |
| RNAseq data | PPMS for EIGC https://fex.cores.emory.edu/ | KYE22363-64287 |
| Experimental Models: Organisms/Strains | ||
| Mouse: 3xTg-AD | Jackson Laboratory | 34 830 |
| Bacteria: Bacteroides fragilis VPI 2553 | ATCC | Cat# 25 285 |
| Bacteria: Bacteroides intestinalis 341 | Leibniz-Institut DSMZ | Cat# DSM 17393 |
| Bacteria: Bacteroides xylanisolvens XB1A | Leibniz-Institut DSMZ | Cat# DSM 18836 |
| Sequence-Based Reagents | ||
| ALOX5 TaqMan Gene Expression Assays | Thermo Fisher | Cat# Hs00167536_m1 |
| PTGS1 TaqMan Gene Expression Assays | Thermo Fisher | Cat# Hs00377721_m1 |
| PTGS2 TaqMan Gene Expression Assays | Thermo Fisher | Cat# Hs00153133_m1 |
| LTB4R TaqMan Gene Expression Assays | Thermo Fisher | Cat# Hs01938704_s1 |
| LTB4R2 TaqMan Gene Expression Assays | Thermo Fisher | Cat# Hs01885851_s1 |
| CEBPB TaqMan Gene Expression Assays | Thermo Fisher | Cat# Mm00843434_s1 |
| GAPDH TaqMan Gene Expression Assays | Thermo Fisher | Cat# Mm99999915_g1 |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Imaris | Oxinst | https://imaris.oxinst.com |
| Prism-GraphPad | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| QIIME | Dr.Alam Ashfaqul laboratory, University of Kentucky | https://qimme.org/ |
| Illumina’s bcl2fastq | Dr.Alam Ashfaqul laboratory, University of Kentucky | https://www.illumina.com/ |
AEP, asparagine endopeptidase; LTB4, leukotriene B4; N/A, not available; QIIME, Quantitative Insights into Microbial Ecology.
Generation of GF mice
3×Tg mice were ordered from the Jackson Laboratory (34830). Animal care and handling were performed according to NIH animal care guidelines and the Declaration of Helsinki and Emory Medical School guidelines. The animals were randomly allocated to experimental groups. Only female mice were used for all the experiments.
GF B6;129-Tg (APPSwe, tauP301L)1Lfa Psen1tm1Mpm/Mmjax (3xTg) were generated in collaboration with Taconic Biosciences (Rensselaer, New York, USA). To create GF 3×Tg-AD mice, at Taconic Biosciences, a 3×Tg embryo was created through in vitro fertilisation and then transplanted into a GF Swiss Webster mouse. Gestating GF Swiss Webster mice were shipped to Emory University in a germ-free transporter and transferred to isolators maintained by the Emory Gnotobiotic Animal Core (EGAC). A colony of GF 3×Tg mice was established by EGAC from the offspring of these mice. GF status of the 3×Tg mice was confirmed by bacterial 16S rDNA PCR assay paired with anaerobic culture testing, undertaken by IDEXX BioAnalytics (Columbia, Missouri, USA), as well agar Gram staining and plating of faecal pellets on Blood agar petri dishes before incubation in aerobic and anaerobic conditions. All the GF 3×Tg mice were housed in individual isolators until 30–32 weeks of age. The sterilisable GF chow diets (Teklad Global 19% Protein Extruded Rodent Diet, 2019S, ENVIGO) contains slightly higher fatty acid content (3.9% omega-6% and 0.4% omega-3 fatty acids) compared with SPF chow diets (PicoLab Rodent Diet 20, 5053*, LabDiet) (2.17% omega-6% and 0.27% omega-3 fatty acids), which was taken into consideration of the potential loss during the sterilisation step.
Human microbiome associated (HMA) mice were created by oral gavage of human microbiome samples to GF 3xTg mice. The HMA 3×Tg mice were housed in Tecniplast IsoCage P-Bioexclusion System cages till 30–32 weeks of age. GF 3×Tg mice supplemented with SCFAs were also housed within Bioexclusion System cages till 30–32 weeks of age. To handle the mice contained in the bioexclusion cages, the cage was saturated in disinfectant and placed in a sterile biosafety hood. Gloves and forceps for mouse handling were sterilised in disinfectant before opening the cage.
Human donor and criteria
The faeces samples from human donors were generously provided by Dr. John P. Haran and Dr. Beth A. McCormick. Human donor selection criteria were as described previously.12 The human faeces samples were collected from nursing home elders who are 65 years of age and lived in one of four nursing home facilities located in central Massachusetts. All the elders had been living at that facility for 1 month and did not have any diarrheal illness or antimicrobial exposure within the preceding 4 weeks. No elders suffered from dysphagia or had a feeding tube. Any elders with antimicrobial exposure or a diarrhoeal illness during the conduct of the study were excluded from this analysis. The information of donors whose samples were applied in this study were shown in online supplemental table 1.
Recolonisation of GF animals with human microbes
Donor samples were received to us frozen at −80°C (after having been collected over time and frozen by collaborators) and allocated into ~0.5 g portions. A single 0.5 g portion was then resuspended in 5 mL sodium bicarbonate (5%). GF 3×Tg mice received 200 uL of the faeces suspension once at 4–5 weeks of age, and colonisation was allowed to take place. A total of 8–10 mice were recolonised with each donor sample. The recipient mice were housed in the separate sterile cages and kept in the biosafety cubical until 30–32 weeks of age.
SCFAs treatment
SCFAs treated GF 3×Tg mice were provided with drinking water containing sodium acetate (67.5 mM; Sigma Aldrich), sodium propionate (25 mM; Sigma Aldrich) and sodium butyrate (40 mM; Sigma Aldrich) beginning at 5–6 weeks of age until 30–32 weeks.
PGE2-G treatment
PGE2-G (Cayman chemicals, 10 mg/mL) was brought to room temperature right before used and diluted with sterilised 1×PBS to 1 mg/mL. GF 3×Tg mice received IP injection of the diluted PGE2-G solution immediately at the dose of 5 mg/kg twice a week for 4 weeks.
Y-Maze behavioural test
All experimental mice in home cage were placed in sterile biosafety hood in the testing room of the SPF grade animal facility for at least 1 hour before testing to minimise effects of stress during testing. The biosafety hood was thoroughly cleaned with Virkon between trials. During the test, experimental mouse was placed just inside arm S facing away from centre and allowed to move through apparatus for a total period of 8 min while being monitored by recording system. Trial begins immediately and ends when defined duration has elapsed. Scoring consists of recording each arm entry (defined as all four paws entering arm). Scoring was conducted live by monitoring movement through apparatus via recorder screen. Experimental mouse was returned to home cage and number of faecal pellets was also recorded. Arena was cleaned with Virkon between trials. Total number of entries did not include first recorded arm (always S). All experimental mice were subjected to Y-maze behavioural test at 30–32 weeks of age.
Number of triads = (Total entries – 2)
*Triad (set of three letters) containing all three letters is scored as alternation
Percent alternation = [(number of Alternations/Total number of triads)×100].
Immunostaining and microglia reconstruction
Cryo-sections of mouse brain were treated with 3% H2O2 for 10 min, followed by three times wash in phosphate buffered saline and 30 min blocking in 1% RIA-BSA,0.3% Triton X-100 as well as the overnight incubation with Iba-1 antibody (1:200) and AEP antibody (11B7) (1:500) at 4°C. The signal was developed using Histostain-SP kit (Invitrogen). To detect the localisation of Aβ, the slides were incubated with Aβ antibody (1:200) at 4°C. To detect the localisation of AT8 and T22, the slides were incubated with AT8 (1:200), T22 (1:700) at 4°C. To detect the localisation of CEBP/β and AEP, the slides were incubated with CEBP/β antibody (H-7) (1:100), AEP antibody (D6S4H) (1:200) at 4°C. To detect the localisation of AEP, AEP-derived tau fragment and phosphorylated Tau in mouse brain section, the slides were incubated with AEP antibody (11B7) (1:500), Tau N368 (homemade,1:1000), APPC586 (homemade, 1:1000) at 4°C. To detect the localisation of CEBP/β, LOX5, COX1, COX2, BLT1, and BLT2, the slides were incubated with CEBP/β antibody (H-7) (1:100), LOX5 antibody (1:200), COX1 antibody (1:200), COX2 antibody (1:200), BLT1 antibody (1:200) and BLT2 antibody (1:200) at 4°C. After overnight incubation, the slides were washed three times in PBS and incubated with Texas Red-conjugated anti-rabbit IgG or FITC-conjugated anti-mouse IgG for 1 hour at room temperature. The slides were washed three times in phosphate buffered saline, then covered with a glass cover using mounting solution and examined under a fluorescence microscope (Olympus). For microglia reconstruction, Z stacks were imaged at 1 μm steps and subsequently analysed using Imaris software.
Aβ plaque staining
Amyloid plaques were stained with Thioflavin-S. The deparaf-finised and hydrated sections were incubated in 0.25% potassium permanganate solution for 20 min, rinsed in distilled water, and incubated in bleaching solution containing 2% oxalic acid and 1% potassium metabisulfite for 2 min. After rinsed in distilled water, the sections were transferred to blocking solution containing 1% sodium hydroxide and 0.9% hydrogen peroxide for 20 min. The sections were incubated for 5 s in 0.25% acidic acid, then washed in distilled water and stained for 5 min with 0.0125% Thioflavin-S in 50% ethanol. The sections were washed with 50% ethanol and placed in distilled water. Then the sections were covered with glass cover using mounting solution.
Western blot analysis
The mice brain tissue was lysed in lysis buffer (50 mM Tris, pH 7.4, 40 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1.5 mM Na VO, 50 mM NaF, 10 mM sodium pyrophosphate, 10 mM sodium β-glycerophosphate, supplemented with protease inhibitors cocktail) and centrifuged for 15 min at 16 000 g. The supernatant was boiled in SDS loading buffer. After SDS-PAGE, the samples were transferred to a nitrocellulose membrane. Western blotting analysis was performed with a variety of antibodies.
AEP activity assay
Tissue homogenates or cell lysates (10 μg) were incubated in 200 μL reaction buffer (20 mM citric acid, 60 mM Na2HPO4, 1 mM EDTA, 0.1% CHAPS and 1 mM DTT, pH 5.5) containing 20 μM AEP substrate Z-Ala-Ala-Asn-AMC (Bachem). AMC released by substrate cleavage was quantified by measuring at 460 nm in a fluorescence plate reader at 37°C in kinetic mode.
RNA sequencing analysis
RNA extraction was performed using the miRNEAsy mini kit (Qiagen) and quantified using the Ribogreen fluorescence protocol on the Infinite M200 Pro (Tecan). Quality of the RNA was evaluated using RNA 6000 Nano reagents on the 2100 bioanalyzer (Agilent). RNA sequencing library preparation was performed using the NEBNext Ultra RNA Library Prep Kit for Illumina by following the manufacturer’s recommendations (NEB). Sequencing libraries were validated on the Agilent 2100 Bioanalyzer System (Agilent Technologies) and quantified using Qubit 2.0 Fluorometer (Invitrogen) as well as by qPCR (Applied Biosystems). The libraries were sequenced on an Illumina sequencer using a 2×150 Paired-End (PE) configuration. Raw sequence data (.bcl files) was converted into fastq files and demultiplexed using Illumina’s bcl2fastq software. Data were aligned and normalised using STAR aligner. Differentially expressed genes were identified by DEseq2. Gene pathway analysis was performed with the database for annotation, visualisation, and integrated discovery (KEGG and PANTHER pathway enrichment analyses). We determined statistically significant differences in gene expression at the nominal level of significance (p<0.05). We also evaluated the effect of Benjamini-Hochberg correction on raw p-values to account for multiple hypothesis testing. The PANTHER (Protein Analysis THrough Evolutionary Relationships) Classification System was used to classify genes. PANTHER Classification System (http://PANTHERdb.org)75 was applied to classify genes that were significantly expressed detected by the DESeq analysis. First, lists of Ensembl gene IDs were recorded in the PANTHER database to classify genes into functional groups, which provided information about their biological function and pathways. Next, the PANTHER statistical over- and underrepresentation tests were performed with log2 fold change (log2FC) values and available Entrez Gene IDs. Finally, it was determined whether identified biological processes were statistically over-or under-expressed. Online supplemental figure 5D contains a Table showing fold enrichment and p values from PANTHER analysis of enriched pathways.
Protocol for microbiota analysis
DNA was extracted from stool samples (n=5 mice per group) using a PowerSoil kit from MO BIO Laboratories (Carlsbad, California, USA). 16S rRNA genes were PCR-amplified from each sample using a composite for- ward primer and a reverse primer containing a unique 12-base barcode, designed using the Golay error-correcting scheme, which was used to tag PCR products from respective samples. We used primers for paired-end 16S community sequencing on the Illumina platform using bacteria/archaeal primer 515F/806R. Primers were specific for the V.4 region of the 16S rRNA gene. The forward PCR primer sequence contained the sequence for the 5′ Illumina adapter, the forward primer pad, the forward primer linker, and the forward primer sequence. Each reverse PCR primer sequence contained the reverse complement of the 3′ Illumina adapter, the Golay barcode (each sequence contained a different barcode), the reverse primer pad, the reverse primer linker, and the reverse primer. Three independent PCR reactions were performed for each sample, combined, and purified with AMPure magnetic purification beads (Agencourt). The products were quantified, and a master DNA pool was generated from the purified products in equimolar ratios. The pooled products were sequenced using an Illumina MiSeq sequencing platform. Bioinformatics analysis was performed using Quantitative Insights into Microbial Ecology. Sequences were assigned to OTUs with UPARSE using 97% pairwise identity and were classified taxonomically using theRibosomal Database Project classifier retrained with Greengenes. After chimaera removal, the average number of reads per sample was 21 511. A single representative sequence for each OTU was aligned using PyNAST, and a phylogenetic tree was then built using FastTree. The phylogenetic tree was used to compute the UniFrac distances. The PCoA analysis shown is unweighted.
Enteric commensal-specific qPCR
For quantification, total microbial DNA and universal bacterial primer sets 515F and 806R were used. To quantify the abundance of specific bacterial species or genus, we used the validated forward and reverse primers specific for the 16S rRNA genes. Detection was achieved using a Fast SYBR Green Master mix according to the manufacturer’s instructions. The following reverse transcription PCR (RT-PCR) protocol was followed: 95°C for 15 min, 40 cycles of 95°C for 15 s, 66°C (bacteria-specific) and 50°C (universal) for 40 s, and 72°C for 30 s. A melting curve was performed after amplification to distinguish between the targeted and nontargeted PCR products. All reactions were performed in duplicate. Bacterial abundance was analysed as genome equivalents. For qPCR analyses, the sample-specific relative abundance of bacteria was determined as genome equivalents amplified by bacteria-specific primers divided by genome equivalents amplified by universal primers. Absolute abundance per gram of stools was determined by adjusting for the dilutions performed during DNA extraction, normalisation, and qPCR setup and dividing this concentration by the total grams of stools used for the original DNA extraction. Last, data were expressed as a fold change in stools collected from different mouse backgrounds compared with stool of the WT mice.
In vitro anaerobic bacteria culture experiment
B. fragilis (25285, ATCC), B. intestinalis 341(DSM17393, DSMZ) and B. xylanisolvens XB1A (DSM18836, DSMZ) were cultured in the chopped meat medium (AS-811, AnaerobeSystems) and incubated at 37°C in Bio-bag with the GasPak EZ Gas Generating Container Systems (260001, Gaspak EZ W/Indicator, BD). For AA metabolism assay, the above bacteria were grown in medium added with or without 62.5 μM AA. 72 hours later, the medium from each culture was collected for the HPLC analysis or ELISA assay.
HPLC quantitative analysis of AA and its metabolites
Solid phase extraction (SPE) sample preparation: Samples were centrifuged for 15 min at 4000 rpm at 4°C to remove bacteria and media pellets. A 10 mL of the supernatant was collected and 10 μL of formic acid and 1 mL of methanol were added to bring the media to contain 10% methanol and 1% formic acid. The samples were sat on ice for at least 15 mins to allow protein precipitation. The samples were centrifuged for 15 min at 4000 rpm at 4°C to remove any precipitations.
As for the control media without inoculating bacteria, the stock standards were added to bring the standard concentration to X50, X100 and X1000 of the stock concentration. The stock concentrations of PGE2, AA, LTB4 and 12HHTrE were 200 μg/mL. 250 μg/mL, 2 μg/mL and 2 μg/mL respectively. The compounds were extracted using SPE columns (Thermo Scientific, CA). Columns were washed with 2 mL of MeOH followed by 2 mL of H2O, 10% methanol with 1% formic acid. 8 mL of the samples or 2 mL of the control medium were added for extraction. After applying the sample, the columns were washed with 4 mL of 10% MeOH, and the analytes were then eluted with 1 mL of MeOH. The eluent was diluted 1:1 with H2O to bring down the methanol to 50% for HPLC assay. The injection volume was 200 μl and 20 μL.
AA and its metabolites, LTB4, 12HHTrE, PGE2 were measured by high-performance liquid chromatography with photodiode array autosampler, model 1525 binary pump and model 2996 photodiode array detector was used. Analytes were separated using reverse-phase chromatography on a Waters Xbridge BEH column (4.6×150 mm, 5 μM, Waters) with guard cartridge (3.9×5 mm, 5 μM, Waters). The analytes were identified by comparing their retention times and spectral profiles to known standards. A stock solution in 100% methanol and the concentrations are 250 μg/mL for AA, 2 μg/mL of LTB4 and 12HHTrE, 200 μg/mL for PGE2, respectively. The working standard solution were prepared in 50% methanol. A series dilution of standard was prepared in 50% methanol as the standard curve.
PGE2 ELISA
Samples were centrifuged for 15 min at 4000 rpm at 4°C to remove bacteria and media pellets. 10 mL of the supernatant was collected and 10 μl of formic acid and 1 mL of methanol were added to bring the media to contain 10% methanol and 1% formic acid. The samples were sat on ice for at least 15 mins to allow protein precipitation. The samples were centrifuged for 15 min at 4000 rpm at 4°C to remove any precipitations. The supernatant was analysed by PGE2 ELISA kit according to the manufacturer’s instructions. The PGE2 concentrations were determined by comparison with the standard curve.
Leukotriene B4 ELISA
Samples were centrifuged for 15 min at 4000 rpm at 4°C to remove bacteria and media pellets. A 10 mL of the supernatant was collected and 10 μL of formic acid and 1 mL of methanol were added to bring the media to contain 10% methanol and 1% formic acid. The samples were sat on ice for at least 15 mins to allow protein precipitation. The samples were centrifuged for 15 min at 4000 rpm at 4°C to remove any precipitations. The supernatant was analysed by LTB4 ELISA kit according to the manufacturer’s instructions. The LTB4 concentrations were determined by comparison with the standard curve.
Golgi staining
Mice brains were fixed in 10% formalin for 24 hours, and then immersed in 3% potassium bichromate for 3 days in the dark. The solution was changed each day. Then the brains were transferred into 2% silver nitrate solution and incubate for 24 hours in the dark. Vibratome sections were cut at 60 μm, air dried for 10 min, dehydrated through 95% and 100% ethanol, cleared in xylene and cover-slipped. For measurement of spine density, only spines that emerged perpendicular to the dendritic shaft were counted.
Aβ ELISA
The mice brains were homogenised in 8X mass of 5 M guanidine HCl/50 mM Tris HCl (pH 8.0), and incubated at room temperature for 3 hours. Then the samples were diluted with cold reaction buffer (phosphate buffered saline with 5% BSA and 0.03% Tween20, supplemented with protease inhibitor cocktail), and centrifuged at 16 000 g for 20 min at 4°C. The supernatant was analysed by human Aβ40 and Aβ42 ELISA kit according to the manufacturer’s instructions. The Aβ concentrations were determined by comparison with the standard curve.
Proinflammatory cytokines ELISA
The mice brain tissue was lysed in lysis buffer (50 mM Tris, pH 7.4, 40 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1.5 mM Na3VO4, 50 mM NaF, 10 mM sodium pyrophosphate, 10 mM sodium β-glycerophosphate, supplemented with protease inhibitors cocktail), and centrifuged for 15 min at 16 000 g. Then the supernatants were analysed by IL-6, TNFα, IL-1β ELISA kit according to the manufacturer’s instructions. The proinflammatory cytokines concentrations were determined by comparison with the standard curve.
Statistical analysis
All data are expressed as mean±SEM from three or more independent experiments, and the level of significance between two groups was assessed with Student’s t-test. For more than two groups, one-way analysis of variance followed by LSD post hoc test was applied. A value of p<0.05 was considered to be statistically significant.
Supplementary Material
Significance of this study.
What is already known on this subject?
The gut microbiome is implicated in various neurological diseases, including Alzheimer’s disease (AD).
Chronic neuroinflammation mediates amyloid-β deposition and neurofibrillary tangles formation, two pathological hallmarks of AD.
C/EBPβ/asparagine endopeptidase (AEP) signalling is temporally activated in the gut of AD patients and 3×Tg mice.
Gut inflammation leads to C/EBPβ/AEP signalling and fibril formation in the gut and brain.
What are the new findings?
Colonisation with gut microbiota from specific-pathogen-free mice and AD patients activate C/EBPβ/AEP signalling in ex-germfree 3×Tg mice, promoting AD pathologies and cognitive deficits.
AD-derived gut microbiota enhance the proinflammatory pathway for poly-unsaturated fatty acid (PUFA) metabolism in the brain.
Bacteroides mediating proinflammatory PUFA metabolism are increased in AD patient gut microbiomes, regulating microglia activation in the brain.
Short chain fatty acids and inflammatory PUFA metabolite stimulate microglia maturation and AD pathologies.
How might it impact on clinical practice in the foreseeable future?
Our findings suggest that the human microbiome could be a risk factor for Alzheimer’s disease. Modulation of gut microbiota through personalised diet or beneficial microbiota intervention, as well as faecal matter transplant may have utility for diminishing the inflammation and treating brain disorders including AD.
Acknowledgements
This study was supported in part by the Emory Gnotobiotic Animal (EGAC), which is subsidised by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the Rodent Behavioral Core (RBC), which is subsidised by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities; the Emory Integrated Genomics Core (EIGC), which is subsidised by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities; as well as Emory HPLC Bioanalytical Core (EHBC), which was supported by the Department of Pharmacology, Emory University School of Medicine. The metabolomics analysis on the feces, serum and brain samples from the AD and HC humanised ex-GF mice and the brains from the GF and SPF 3xTg mice was performed by Metabolon, Morrisville, North Carolina, USA.
Funding
This work is supported by a grant from the National Institute of Health (RO1, AG065177) to KY. Additional support was provided by the Georgia Clinical and Translational Science Alliance of the National Institutes of Health under award number UL1TR002378 and Emory ADRC grant P30 AG066511.
Footnotes
Competing interests None declared.
Patient consent for publication Not applicable.
Ethics approval The experimental protocol was approved by the Emory University Institutional Animal Care and Ethical Committee (IACUC).
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information.
REFERENCES
- 1.Heneka MT, Carson MJ, El Khoury J, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol 2015;14:388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wendeln A-C, Degenhardt K, Kaurani L, et al. Innate immune memory in the brain shapes neurological disease hallmarks. Nature 2018;556:332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cagnin A, Brooks DJ, Kennedy AM, et al. In-Vivo measurement of activated microglia in dementia. Lancet 2001;358:461–7. [DOI] [PubMed] [Google Scholar]
- 4.Griciuc A, Serrano-Pozo A, Parrado AR, et al. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 2013;78:631–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz Heijtz R, Wang S, Anuar F, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 2011;108:3047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 2012;10:735–42. [DOI] [PubMed] [Google Scholar]
- 7.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature 2007;449:819–26. [DOI] [PubMed] [Google Scholar]
- 8.Cattaneo A, Cattane N, Galluzzi S, et al. Association of brain amyloidosis with proinflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging 2017;49:60–8. [DOI] [PubMed] [Google Scholar]
- 9.Liu P, Wu L, Peng G, et al. Altered microbiomes distinguish Alzheimer’s disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav Immun 2019;80:633–43. [DOI] [PubMed] [Google Scholar]
- 10.Vogt NM, Kerby RL, Dill-McFarland KA, et al. Gut microbiome alterations in Alzheimer’s disease. Sci Rep 2017;7:13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang Z-Q, Shen L-L, Li W-W, et al. Gut microbiota is altered in patients with Alzheimer’s disease. J Alzheimers Dis 2018;63:1337–46. [DOI] [PubMed] [Google Scholar]
- 12.Haran JP, Bhattarai SK, Foley SE, et al. Alzheimer’s disease microbiome is associated with dysregulation of the anti-inflammatory P-glycoprotein pathway. mBio 2019;10:e00632–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandscheid C, Schuck F, Reinhardt S, et al. Altered gut microbiome composition and tryptic activity of the 5XFAD Alzheimer’s mouse model. J Alzheimers Dis 2017;56:775–88. [DOI] [PubMed] [Google Scholar]
- 14.Bäuerl C, Collado MC, Diaz Cuevas A, et al. Shifts in gut microbiota composition in an APP/PSS1 transgenic mouse model of Alzheimer’s disease during lifespan. Lett Appl Microbiol 2018;66:464–71. [DOI] [PubMed] [Google Scholar]
- 15.Chen C, Ahn EH, Kang SS, et al. Gut dysbiosis contributes to amyloid pathology, associated with C/EBPβ/AEP signaling activation in Alzheimer’s disease mouse model. Sci Adv 2020;6:eaba0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harach T, Marungruang N, Duthilleul N, et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci Rep 2017;7:41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farooqui AA, Horrocks LA, Farooqui T. Modulation of inflammation in brain: a matter of fat. J Neurochem 2007;101:577–99. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Mejia RO, Mucke L. Phospholipase A2 and arachidonic acid in Alzheimer’s disease. Biochim Biophys Acta 2010;1801:784–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi S-H, Bosetti F. Cyclooxygenase-1 null mice show reduced neuroinflammation in response to beta-amyloid. Aging 2009;1:234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinne JO, Laakso K, Lönnberg P, et al. Brain muscarinic receptors in senile dementia. Brain Res 1985;336:19–25. [DOI] [PubMed] [Google Scholar]
- 21.Esposito G, Giovacchini G, Liow J-S, et al. Imaging neuroinflammation in Alzheimer’s disease with radiolabeled arachidonic acid and PET. J Nucl Med 2008;49:1414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Mejia RO, Newman JW, Toh S, et al. Phospholipase A2 reduction ameliorates cognitive deficits in a mouse model of Alzheimer’s disease. Nat Neurosci 2008;11:1311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Chen C. Cyclooxygenase-2 in synaptic signaling. Curr Pharm Des 2008;14:1443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amtul Z, Uhrig M, Wang L, et al. Detrimental effects of arachidonic acid and its metabolites in cellular and mouse models of Alzheimer’s disease: structural insight. Neurobiol Aging 2012;33:831.e21–831.e31. [DOI] [PubMed] [Google Scholar]
- 25.Smirnov A, Trupp A, Henkel AW, et al. Differential processing and secretion of Abeta peptides and sAPPalpha in human platelets is regulated by thrombin and prostaglandine 2. Neurobiol Aging 2009;30:1552–62. [DOI] [PubMed] [Google Scholar]
- 26.Firuzi O, Zhuo J, Chinnici CM, et al. 5-Lipoxygenase gene disruption reduces amyloid-beta pathology in a mouse model of Alzheimer’s disease. Faseb J 2008;22:1169–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannopoulos PF, Chu J, Joshi YB, et al. 5-Lipoxygenase activating protein reduction ameliorates cognitive deficit, synaptic dysfunction, and neuropathology in a mouse model of Alzheimer’s disease. Biol Psychiatry 2013;74:348–56. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Chu J, Giannopoulos PF, Ceballos-Diaz C, et al. 5-Lipoxygenase gene transfer worsens memory, amyloid, and tau brain pathologies in a mouse model of Alzheimer disease. Ann Neurol 2012;72:442–54. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Zhang Z, Song M, Liu X, et al. Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer’s disease. Nat Med 2014;20:1254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Song M, Liu X, et al. Delta-secretase cleaves amyloid precursor protein and regulates the pathogenesis in Alzheimer’s disease. Nat Commun 2015;6:8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z-H, Gong K, Liu X, et al. C/Ebpβ regulates delta-secretase expression and mediates pathogenesis in mouse models of Alzheimer’s disease. Nat Commun 2018;9:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strohmeyer R, Shelton J, Lougheed C, et al. Ccaat-Enhancer binding protein-β expression and elevation in Alzheimer’s disease and microglial cell cultures. PLoS One 2014;9:e86617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ndoja A, Reja R, Lee S-H, et al. Ubiquitin ligase COP1 suppresses neuroinflammation by degrading C/EBPβ in microglia. Cell 2020;182:1156–69. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Zhou Y, Wang H, et al. Gut inflammation triggers C/EBPβ/δ-secretase- dependent gut-to-brain propagation of Aβ and tau fibrils in Alzheimer’s disease. Embo J 2021;40:e106320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Liu X, Chen S, et al. Spatiotemporal activation of the C/EBPβ/δ-secretase axis regulates the pathogenesis of Alzheimer’s disease. Proc Natl Acad Sci U S A 2018;115:E12427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang Y-J, Mbonye UR, DeLong CJ, et al. Regulation of intracellular cyclooxygenase levels by gene transcription and protein degradation. Prog Lipid Res 2007;46:108–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talbot K, Wang H-Y, Kazi H, et al. Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J Clin Invest 2012;122:1316–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steen E, Terry BM, Rivera EJ, et al. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease--is this type 3 diabetes? J Alzheimers Dis 2005;7:63–80. [DOI] [PubMed] [Google Scholar]
- 39.Schafer DP, Stevens B. Microglia function in central nervous system development and plasticity. Cold Spring Harb Perspect Biol 2015;7:a020545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalsotra A, Anakk S, Brommer CL, et al. Catalytic characterization and cytokine mediated regulation of cytochrome P450 4Fs in rat hepatocytes. Arch Biochem Biophys 2007;461:104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu P, Fleete MS, Jing Y, et al. Altered arginine metabolism in Alzheimer’s disease brains. Neurobiol Aging 2014;35:1992–2003. [DOI] [PubMed] [Google Scholar]
- 42.Straccia M, Dentesano G, Valente T, et al. Ccaat/Enhancer binding protein β regulates prostaglandin E synthase expression and prostaglandin E2 production in activated microglial cells. Glia 2013;61:1607–19. [DOI] [PubMed] [Google Scholar]
- 43.Reddy KV, Serio KJ, Hodulik CR, et al. 5-Lipoxygenase-Activating protein gene expression. key role of CCAAT/enhancer-binding proteins (C/EBP) in constitutive and tumor necrosis factor (TNF) alpha-induced expression in THP-1 cells. J Biol Chem 2003;278:13810–8. [DOI] [PubMed] [Google Scholar]
- 44.Serio KJ, Reddy KV, Bigby TD. Lipopolysaccharide induces 5-lipoxygenase-activating protein gene expression in THP-1 cells via a NF-kappaB and C/EBP-mediated mechanism. Am J Physiol Cell Physiol 2005;288:C1125–33. [DOI] [PubMed] [Google Scholar]
- 45.Healy ZR, Zhu F, Stull JD, et al. Elucidation of the signaling network of COX-2 induction in sheared chondrocytes: COX-2 is induced via a Rac/MEKK1/MKK7/JNK2/c-Jun-C/EBPbeta-dependent pathway. Am J Physiol Cell Physiol 2008;294:C1146–57. [DOI] [PubMed] [Google Scholar]
- 46.Park JY, Pillinger MH, Abramson SB. Prostaglandin E2 synthesis and secretion: the role of PGE2 synthases. Clin Immunol 2006;119:229–40. [DOI] [PubMed] [Google Scholar]
- 47.Murakami M, Nakashima K, Kamei D, et al. Cellular prostaglandin E2 production by membrane-bound prostaglandin E synthase-2 via both cyclooxygenases-1 and −2. J Biol Chem 2003;278:37937–47. [DOI] [PubMed] [Google Scholar]
- 48.Conteh AM, Reissaus CA, Hernandez-Perez M, et al. Platelet-Type 12-lipoxygenase deletion provokes a compensatory 12/15-lipoxygenase increase that exacerbates oxidative stress in mouse islet β cells. J Biol Chem 2019;294:6612–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Straccia M, Gresa-Arribas N, Dentesano G, et al. Pro-Inflammatory gene expression and neurotoxic effects of activated microglia are attenuated by absence of CCAAT/enhancer binding protein β. J Neuroinflammation 2011;8:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Z, Liu X, Cheng L, et al. Delta-secretase triggers Alzheimer’s disease pathologies in wild-type hAPP/hMAPT double transgenic mice. Cell Death Dis 2020;11:1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortes-Canteli M, Luna-Medina R, Sanz-Sancristobal M, et al. Ccaat/Enhancer binding protein beta deficiency provides cerebral protection following excitotoxic injury. J Cell Sci 2008;121:1224–34. [DOI] [PubMed] [Google Scholar]
- 52.Ikeda-Matsuo Y, Hirayama Y, Ota A, et al. Microsomal prostaglandin E synthase-1 and cyclooxygenase-2 are both required for ischaemic excitotoxicity. Br J Pharmacol 2010;159:1174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakayama M, Uchimura K, Zhu RL, et al. Cyclooxygenase-2 inhibition prevents delayed death of CA1 hippocampal neurons following global ischemia. Proc Natl Acad Sci U S A 1998;95:10954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puzzo D, Lee L, Palmeri A, et al. Behavioral assays with mouse models of Alzheimer’s disease: practical considerations and guidelines. Biochem Pharmacol 2014;88:450–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandal M, White PJ, Chevrier G, et al. Age-Dependent impairment of glucose tolerance in the 3xTg-AD mouse model of Alzheimer’s disease. Faseb J 2015;29:4273–84. [DOI] [PubMed] [Google Scholar]
- 56.Yu Y, Li X, Blanchard J, et al. Insulin sensitizers improve learning and attenuate tau hyperphosphorylation and neuroinflammation in 3xTg-AD mice. J Neural Transm 2015;122:593–606. [DOI] [PubMed] [Google Scholar]
- 57.Du K, Ding J. Insulin regulates TRB3 and other stress-responsive gene expression through induction of C/EBPbeta. Mol Endocrinol 2009;23:475–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foti D, Iuliano R, Chiefari E, et al. A nucleoprotein complex containing Sp1, C/EBP beta, and HMGI-Y controls human insulin receptor gene transcription. Mol Cell Biol 2003;23:2720–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Y, Xiong K, Shen M, et al. CCAAT-Enhancer binding protein (C/EBP) β regulates insulin-like growth factor (IGF) 1 expression in porcine liver during prenatal and postnatal development. Mol Cell Biochem 2015;401:209–18. [DOI] [PubMed] [Google Scholar]
- 60.Chen DY, Stern SA, Garcia-Osta A, et al. A critical role for IGF-II in memory consolidation and enhancement. Nature 2011;469:491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarthy TL, Ji C, Chen Y, et al. Time- and dose-related interactions between glucocorticoid and cyclic adenosine 3’,5’-monophosphate on CCAAT/enhancer-binding protein-dependent insulin-like growth factor I expression by osteoblasts. Endocrinology 2000;141:127–37. [DOI] [PubMed] [Google Scholar]
- 62.Chang W, Rewari A, Centrella M, et al. Fos-Related antigen 2 controls protein kinase A-induced CCAAT/enhancer-binding protein beta expression in osteoblasts. J Biol Chem 2004;279:42438–44. [DOI] [PubMed] [Google Scholar]
- 63.Chaudhry UA, Zhuang H, Crain BJ, et al. Elevated microsomal prostaglandin-E synthase-1 in Alzheimer’s disease. Alzheimers Dement 2008;4:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Braniste V, Al-Asmakh M, Kowal C, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med 2014;6:263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mapstone M, Cheema AK, Fiandaca MS, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med 2014;20:415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salek RM, Xia J, Innes A, et al. A metabolomic study of the CRND8 transgenic mouse model of Alzheimer’s disease. Neurochem Int 2010;56:937–47. [DOI] [PubMed] [Google Scholar]
- 67.Inoue K, Tsutsui H, Akatsu H, et al. Metabolic profiling of Alzheimer’s disease brains. Sci Rep 2013;3:2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pan X, Kaminga AC, Wen SW, et al. Dopamine and dopamine receptors in Alzheimer’s disease: a systematic review and network meta-analysis. Front Aging Neurosci 2019;11:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Basen M, Kurrer SE. A close look at pentose metabolism of gut bacteria. Febs J 2021;288:1804–8. [DOI] [PubMed] [Google Scholar]
- 70.Adesso S, Magnus T, Cuzzocrea S, et al. Indoxyl sulfate affects glial function increasing oxidative stress and neuroinflammation in chronic kidney disease: interaction between astrocytes and microglia. Front Pharmacol 2017;8:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Erny D, Hrabě de Angelis AL, Jaitin D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 2015;18:965–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matcovitch-Natan O, Winter DR, Giladi A, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 2016;353:aad8670. [DOI] [PubMed] [Google Scholar]
- 73.Dodiya HB, Kuntz T, Shaik SM, et al. Sex-Specific effects of microbiome perturbations on cerebral Aβ amyloidosis and microglia phenotypes. J Exp Med 2019;216:1542–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rivière A, Selak M, Lantin D, et al. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol 2016;7:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mi H, Muruganujan A, Huang X, et al. Protocol update for large-scale genome and gene function analysis with the Panther classification system (v.14.0). Nat Protoc 2019;14:703–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information.