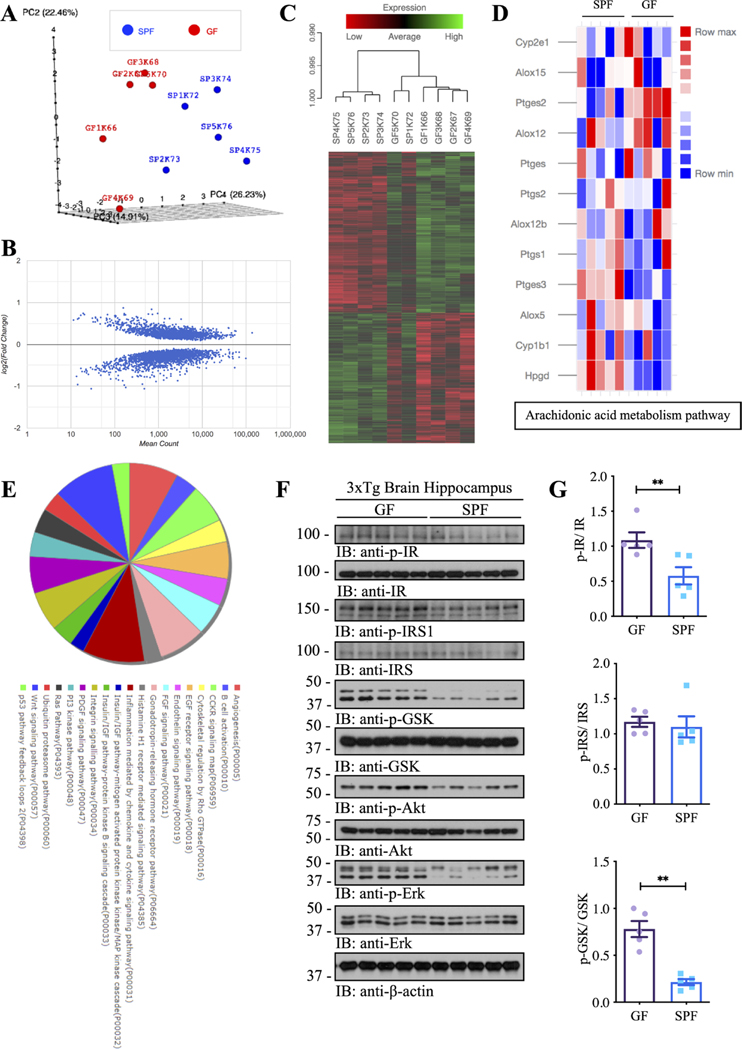
Figure 3.
Gutmicrobiota impacts the transcriptome atlas of mRNA expression in the hippocampal regions of germ-free and SPF 3xTg mouse model of AD. (A) Plot showing sample-to-sample distances in a principal components analysis (PCA). (B) Scatter plot showing log2 (fold change) against normalised mean gene count. (C) Cluster analysis of functional mRNAs in GF and SPF, and data are presented as a heat MAP (p<0.05). Each row represents the relative levels of expression of a single gene across all mice; each column represents the expression levels for a single mouse. The colours red and green denote low and high expression, respectively. (D) Heatmap showing differential genes of Arachidonic acid metabolism pathways between GF and SPF. (E) KEGG and Panther pathway analysis of differential genes. (F) Immunoblot blot showing insulin/IGF-1 signalling pathways in the hippocampus of germ-free 3xTg mice and SPF 3xTg mice. (G) Quantitative analysis of immunoblot. The bands of p-IR, p-IRS and p-GSK were measured with image J and normalised with IR, IRS and GSK, respectively. N=5 in each group, data are shown as mean±SEM, **p<0.01 compared with control, unpaired t-tests. AD, Alzheimer’s disease; GF, germ-free; IR, insulin receptors; SPF, specific-pathogen-free.