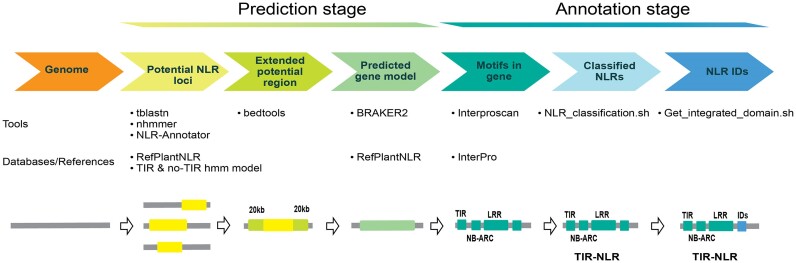
Figure 4:
Workflow of the FindPlantNLRs pipeline: a tool for annotating nucleotide binding and leucine-rich repeat (NLR) genes. The pipeline annotates predicted NLR genes from an unmasked genome fasta file input. We combine loci identified using NLR annotator software with a basic local alignment search tool (tblastn) using recently compiled and functionally validated NLR amino acid sequences and a nucleotide iterative hidden Markov model (HMM) to locate NBARC domains in genomes. The loci identified (including 20-kb flanking regions) are then annotated with Braker2 software using protein hints from experimentally validated resistance genes. Annotated amino acid fasta files are screened for domains using Interproscan, and the predicted coding and amino acid sequences containing both NB-ARC and LRR domains are located back to scaffolds and extracted in gff3 format.