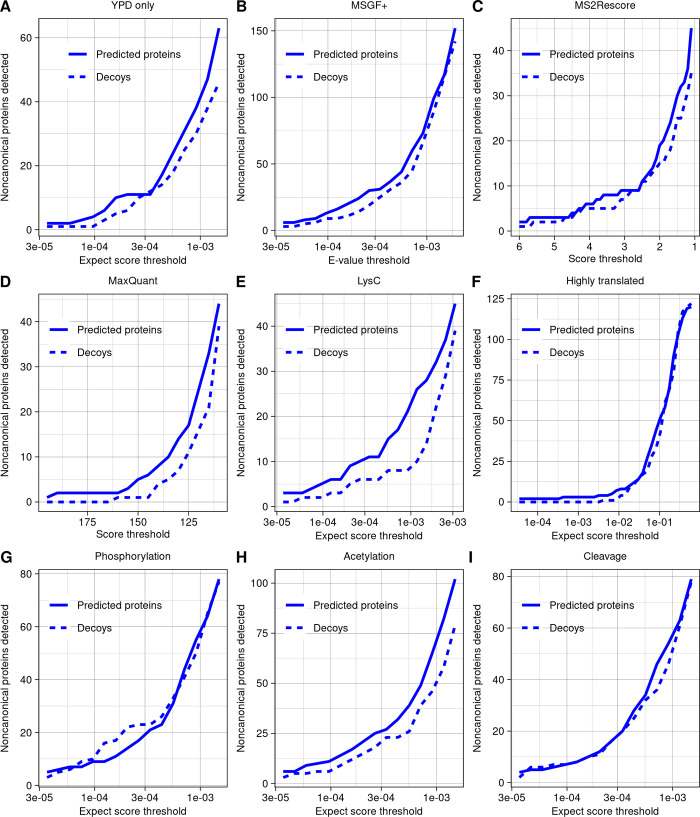
Fig 4. Alternative strategies for detecting noncanonical ORF products yield few additional discoveries.
(A-I) The number of predicted proteins and decoys detected across a range of thresholds, using a variety of strategies for detection. Aside from the specific changes indicated, all searches were run using the same parameter settings. The data underlying this Figure can be found in S1 Data. (A) Analysis using only ribo-seq and MS data taken from yeast grown in YPD at 30°C. (B) Analysis using the program MSGF+. (C) Analysis using the rescoring algorithm MS2Rescore on MSGF+ results. Higher scores indicate higher confidence. (D) Analysis using the program MaxQuant. Higher scores indicate higher confidence. (E) Analysis including only experiments using LysC as protease. (F) Analysis restricted to database of 379 predicted noncanonical proteins encoded by ORFs in the top 2% of in-frame ribo-seq reads per codon. (G) Analysis allowing for phosphorylation of threonine, serine, or tyrosine as variable modifications. (H) Analysis allowing for acetylation of lysine or n-terminal acetylation as variable modifications. (I) Analysis allowing detection of peptides with one end as a non-enzymatic cut site.