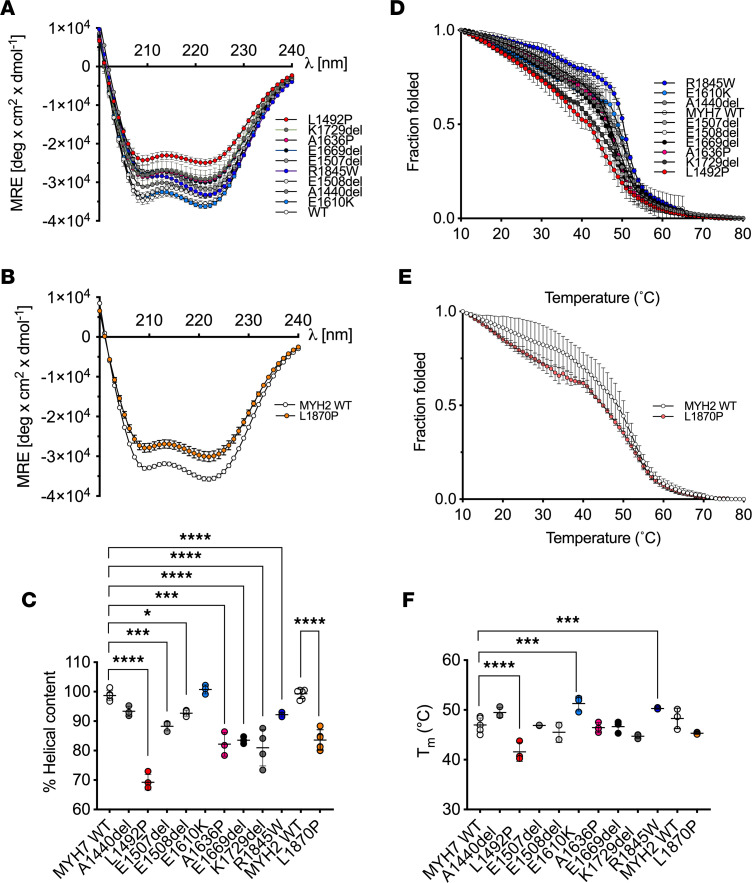
Figure 2. Secondary structure of LMM constructs.
(A and B) The mean circular dichroism (CD) spectra at 10°C for each of the MYH7 (A) and MYH2 (B) LMM constructs. Data are shown as mean ± SD. MRE, mean residue ellipticity. Mutations to proline are shown in shades of red, deletion mutations in shades of gray, and other mutations in shades of blue. (C) The percentage of helicity for each MYH7 and MYH2 LMM construct calculated from the 222 nm MRE values from a minimum of 3 separate experiments. Individual values together with the mean ± SD are shown. The normalized MRE measured at 222 nm from 10°C to 70°C for MYH7 (D) and MYH2 (E) constructs for a minimum of 3 experiments. These data were used to calculate Tm (temperature at which half the protein is melted) for each construct plotted in F. Individual data points for each measurement, together with the mean values ± SD, are shown. Significant differences compared with WT are indicated; the 1-way ANOVA with Dunnett’s test post hoc correction was used with *P < 0.05; ***P < 0.001; ****P < 0.0001. Heptad positions for the mutated residues in MYH7: A1440 and A1636 ‘b’; E1610 and K1729 ‘c’; L1492 ‘e’; E1507 and R1845 ‘f’; E1508 ‘g’ and in MYH2: L1870 ‘d’.