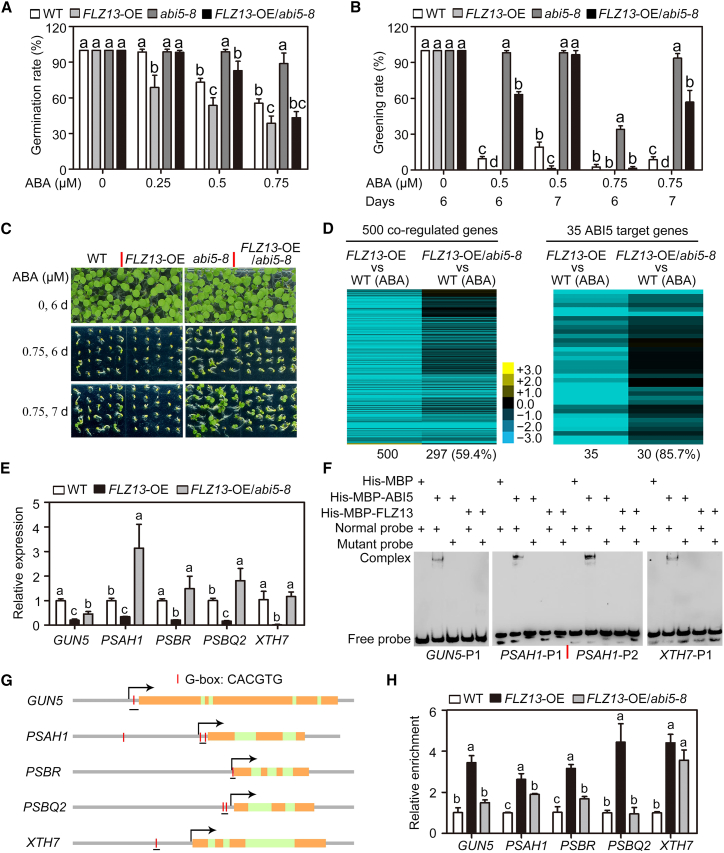
Figure 5.
FLZ13 regulates ABA response during seed germination in an ABI5-dependent manner.
(A) Germination rates of WT, FLZ13-OE, abi5-8, and FLZ13-OE/abi5-8 treated with the indicated concentrations of ABA. Germination rates were recorded 4 days after plating on ½ MS medium. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(B) Cotyledon greening rates of WT, FLZ13-OE, abi5-8, and FLZ13-OE/abi5-8 treated with various concentrations of ABA at different time points, as indicated. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(C) Photographs of WT, FLZ13-OE, abi5-8, and FLZ13-OE/abi5-8 seedlings germinated on medium containing various concentrations of ABA for the indicated time periods.
(D) Heatmap showing the expression of genes synchronously co-regulated by ABA/ABI5/FLZ13 in FLZ13-OE and FLZ13-OE/abi5-8 lines in the presence of ABA. Three-day-old germinating seeds with 0.5 μM ABA treatment were collected for the RNA-seq assay.
(E) Quantitative RT–PCR assay showing expression of the selected genes in WT, FLZ13-OE, and FLZ13-OE/abi5-8 upon ABA treatment. Three-day-old germinating seeds with 0.5 μM ABA treatment were collected for the qRT–PCR assay. PP2A was used as an internal control. Data are presented as mean ± SD (n = 3 technical replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05). This experiment was repeated twice with similar results.
(F) EMSA showing the binding ability of ABI5 and FLZ13 to the ABI5 target genes GUN5, PSAH1, and XTH7. The DNA probes used in this experiment are shown in Supplemental Figure 9. In the mutant probes, the G-box sequence (CACGTG) was replaced with CTTTTG. The probe sequences are listed in Supplemental Table 14.
(G) Structure of selected ABI5 target genes. The positions of the G-box and fragments used for the ChIP–PCR assay are marked.
(H) ChIP–qPCR assay showing the relative enrichment of FLZ13 at the selected gene locus. Data are presented as mean ± SD (n = 3 technical replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05). This experiment was repeated twice with similar results.