Abstract
Three sequences similar to that of the consensus binding sequence of the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex were found in the major IS2 promoter region. Experiments were performed to determine whether the cAMP-CRP complex plays a role in the regulation of IS2 transposition. In the gel retardation assay, the cAMP-CRP complex was found to be able to bind the major IS2 promoter. A DNA footprinting assay confirmed that the cAMP-CRP complex binds to the sequences mentioned above. With an IS2 promoter-luciferase gene fusion construct, the cAMP-CRP complex was shown to inhibit transcription from the major IS2 promoter. IS2 was found to transpose at a frequency approximately 200-fold higher in an Escherichia coli host defective for CRP or adenyl cyclase than in a wild-type host. These results suggest that the cAMP-CRP complex is a negative regulator of IS2 transposition.
The insertion sequence IS2 is a member of the IS3 family (56, 65, 66). It is 1,331 bp in length, with a pair of 42-bp imperfect inverted repeats (18, 26, 64). The IS2 genome contains five open reading frames (ORF1 to -5) of greater than 50 amino acids; however, only two IS2-encoded proteins, of 14 and 46 kDa, have been detected (27, 28). The 14-kDa protein, referred to as the InsA protein (27), is encoded by ORF1. The 46-kDa protein is designated InsAB′. It is encoded by ORF1 and ORF2 via a −1 frameshift mechanism (28) at the frameshift signal AAAAAAG, which is located between the 3′ end of ORF1 and the 5′ end of ORF2 (8, 28, 56). The mRNAs encoding both proteins are transcribed from the promoter located within the left inverted repeat (LIR) of IS2. This promoter has been shown to be the major promoter of IS2 (27). The production of InsAB′ by a −1 frameshift mechanism appears to be a general phenomenon in members of the IS3 family, because it also occurs in IS150 (56, 78), IS911 (55, 56), and IS3 (69).
The 14-kDa InsA is a DNA binding protein. It binds to the sequence 5′-TATCACTTAAATAAGTGATA-3′ (27), which is located around the −10 sequences of the major IS2 promoter (Fig. 1). Since this promoter is responsible for the expression of both InsA and InsAB′, binding of InsA to this sequence may affect transcription. This notion is supported by the observation of a decrease in IS2 transposition when InsA is overexpressed (27). InsA functions as a homodimer. Dimerization of InsA takes place at the C-terminal end of the molecule, whereas the DNA binding domain of InsA is located at its N terminus (38).
FIG. 1.
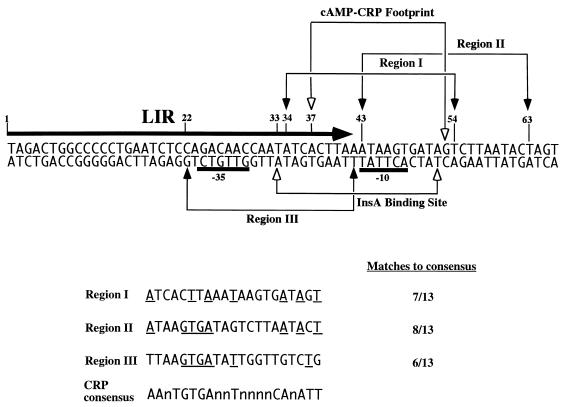
Locations of binding sequences for the cAMP-CRP complex in the major IS2 promoter. The LIR of IS2 is indicated by a large arrow above the sequence. The solid bars below the sequence indicate the locations of the −10 and −35 sequences of the major IS2 promoter. At the bottom, three putative cAMP-CRP complex binding sequences, region I (IS2 nucleotide positions 34 to 54), region II (positions 43 to 63), and region III (positions 42 to 22), present in the promoter area are aligned with the consensus cAMP-CRP complex binding sequence (matching bases are underlined). The cAMP-CRP complex binding sequence (positions 37 to 53) determined by the DNA footprinting experiment is bracketed above the sequence, and the InsA binding site (positions 33 to 52) is indicated below the sequence.
The 46-kDa InsAB′ protein has typical transposase motifs WxxD (36), N3 (61), and C1 (41, 60), collectively known as the DDE motif (20, 36, 54), located at its C terminus (28) and a helix-turn-helix DNA binding motif, TVSLVARQHGVAASQLFLWR, located at its N terminus (amino acid positions 31 to 50) (38). InsAB′ has the ability to bind both terminal repeats of IS2 (28). Overexpression of InsAB′ has been shown to increase IS2 transpositional recombination and formation of two transpositional products, IS2 minicircles and figure eight molecules (39). These observations suggest that InsAB′ is a transposase of IS2.
It is possible that host factors may affect IS2 transposition. Examination of the nucleotide sequence of the IS2 promoter revealed three putative cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex binding sequences located in the major IS2 promoter (Fig. 1). Since cAMP-CRP is a global transcriptional regulator which may activate or inactivate gene expression (34, 50, 62, 63), we investigated the possible role of cAMP-CRP in the regulation of IS2 transposition.
MATERIALS AND METHODS
PCR.
PCRs were performed in a 100-μl mixture containing 10 ng of template DNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100), 20 pmol of each PCR primer, 0.2 mM (each) deoxynucleoside triphosphate, and 2 U of TaqI DNA polymerase. Temperature cycling for PCR included 1 cycle of 95°C for 5 min; 25 cycles of 95°C for 1 min, 48°C for 1 min, and 72.5°C for 2 min; and a 10-min extension at 72.5°C. The PCR products were electrophoresed on a 1.0% agarose gel to determine the sizes of the amplified products.
Gel retardation assay.
Cell lysates of Escherichia coli JM109(DE3) (73) containing pSK−CRP (Fig. 2A), pT7-7 (74), or pT7insA (27) were used as the sources of DNA binding proteins in the gel retardation assay. These cell lysates were prepared by repeated freezing-thawing and sonication of cells from rifampin-treated cultures as described previously (27, 38). Aliquots of clarified cell lysates containing various concentrations of protein were used. The gel retardation reaction mixture contained the cell lysate, a 32P-labeled DNA fragment, 50 mM Tris-HCl (pH 7.4), 70 mM KCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 7 mM MgCl2, 3 mM CaCl2, 10% glycerol, 25 μg of herring sperm DNA, and 200 μg of bovine serum albumin/ml in a total volume of 25 μl. The reaction mixture was incubated at room temperature for 25 min. After the addition of 5 μl of a DNA electrophoresis loading buffer (1 μg of bovine serum albumin/ml, 50% glycerol, 0.01% xylene cyanol) to each reaction mixture, the mixtures were electrophoresed on a 5% native polyacrylamide gel (16). Retarded protein-DNA complex bands were visualized by autoradiography of the gel.
FIG. 2.
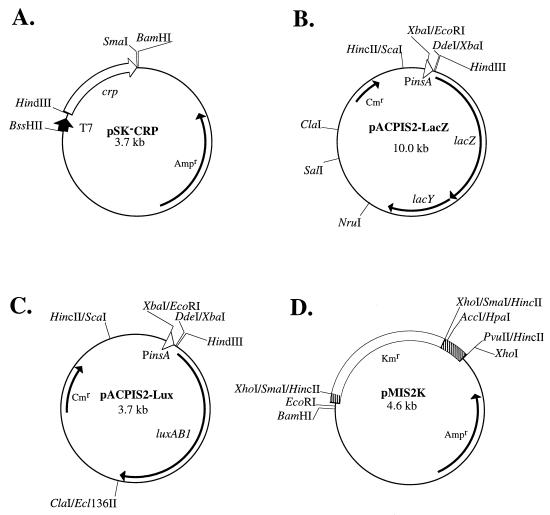
Plasmids used in this study. The shaded regions in panel D are IS2 sequences which remained. Abbreviations: crp, CRP gene; Ampr, ampicillin resistance gene; Cmr, chloramphenicol resistance gene; Kmr, kanamycin resistance gene; luxAB1, luciferase gene from Vibrio harveyi (45); PinsA, the insA gene promoter, which is also the major IS2 promoter.
In situ DNA footprinting.
The DNA-protein reaction mixtures described above were electrophoresed on a 5% native acrylamide gel. After being washed with 200 ml of 50 mM Tris-HCl (pH 8.0) solution, the whole gel was soaked at room temperature for 8 min in an in situ DNA footprinting solution containing 1 mM 1,10-phenanthroline, 0.225 mM CuSO4, and 29 mM 3-mercaptopropionic acid as described previously (27, 37). The gel was washed with water and then exposed to an X-ray film to detect protein-bound DNA bands. The gel was then aligned with the autoradiogram, and the portions of the gel containing the bands were isolated. The DNA present in the gel slices was eluted by soaking the gel in 0.5 ml of a solution of 0.5 M ammonium acetate and 1 mM EDTA overnight. The eluted DNA was precipitated with ethanol and then electrophoresed on a 6% DNA sequencing gel.
Luciferase assay.
One hundred microliters of 1% (vol/vol) n-decyl-aldehyde (in ethanol) was added to 500 μl of a late-log-phase (A600 = 0.8) culture of E. coli containing appropriate plasmids. The reaction mixture was incubated at room temperature for 10 s, and then the luciferase activity was measured with a luminometer (AutoLumat LB 593; EG & G, BERTHOLD, Bad Wildbad, Germany). The bioluminescence generated from each culture was shown as relative light units (RLU).
Transposition assay.
Transposition assays and the determination of transposition frequencies were performed as described previously (26). A kanamycin resistance (Kmr) gene was inserted into IS2 so that transposition of IS2 could be detected by determining kanamycin resistance. pMIS2K (Fig. 2D), which carries this kanamycin gene-marked IS2, was introduced into the isogenic E. coli strains TP7811 (xyl araH1 his), TP7839 (xyl araH1 his Δcrp39), and TP7860 (xyl araH1 his Δcya) (4) containing an F-derived plasmid, pCJ105, which served as the target for IS2 transposition. These three isogenic E. coli strains also harbor IS2, which provides the IS2 transposase in the transposition assay. Since pCJ105 carries a chloramphenicol resistance gene, transposition of IS2 onto pCJ105 will render pCJ105 able to confer on an E. coli host both kanamycin- and chloramphenicol-resistant phenotypes. To determine the transposition frequency, pCJ105::IS2 was mated out from TP7811, TP7839, or TP7860 to HB101 (5) by conjugation. The transconjugants were selected on Luria-Bertani agar containing chloramphenicol (50 μg/ml), kanamycin (50 μg/ml), and streptomycin (50 μg/ml), since HB101 is resistant to streptomycin. The transposition frequency was calculated by dividing the number of HB101 cells that were resistant to kanamycin, chloramphenicol, and streptomycin by those that were resistant to only chloramphenicol and streptomycin.
RESULTS
Binding of cAMP-CRP to the major IS2 promoter.
To determine whether the cAMP-CRP complex has the ability to bind the major IS2 promoter, the gel retardation assay was performed. A cell lysate containing overexpressed CRP was used as the source of CRP for this experiment. To overexpress CRP, a 0.7-kb DNA fragment containing the crp gene was amplified by PCR with genomic DNA isolated from E. coli XL1-Blue (7) as the template and primers CRP-N (5′-TTATCTGGCTCTGGAGAAAGCTT-3′), which has a HindIII site at its 3′ end, and CRP-C (5′-TCGAAGTGCATAGTTGATATCGG-3′). The PCR product was digested with HindIII and then cloned between the HindIII and SmaI sites of pBluescript II SK(−), generating pSK−CRP (Fig. 2A). The nucleotide sequence of the cloned crp gene was verified by sequencing. pSK−CRP was then introduced into E. coli JM109(DE3), and the lysate of JM109(DE3) cells containing pSK−CRP was used for the gel retardation assay.
A DNA fragment referred to as PinsA, which is the 89-bp EcoRI-SpeI fragment containing the IS2 LIR, was isolated from pKS+IS2 (28), labeled at its 3′ end with [α-32P]dATP, and then incubated with cell lysate of JM109(DE3) containing pSK−CRP in the presence or absence of cAMP. The reaction products were then electrophoresed on a polyacrylamide gel to detect bands that migrated more slowly than those in control reactions which lacked CRP or cAMP. The same labeled fragment was reacted with the InsA protein to serve as a positive DNA binding control, since InsA is known to bind the LIR. Binding of cAMP-CRP to the lacZ gene promoter was also performed to serve as an additional positive DNA binding control, because the cAMP-CRP complex binds to the lacZ gene promoter. Reaction of cAMP-CRP with a 170-bp EcoRI-PvuII fragment of pBluescript II SK(+), which does not contain a cAMP-CRP binding site, was done to serve as a negative control.
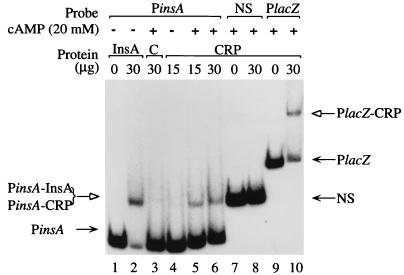
The results of this experiment are shown in Fig. 3. A retarded band which migrated more slowly than the naked PinsA (Fig. 3, lane 1) was seen when PinsA was reacted with the cell lysate (30 μg of total protein) containing InsA (Fig. 3, lane 2), indicating that PinsA has the InsA binding sequence. In the presence of 20 mM cAMP, both 30 and 15 μg of total protein of the cell lysate of JM109(DE3) cells containing pSK−CRP generated a retarded band when incubated with PinsA (Fig. 3, lanes 5 and 6). This retarded band was not seen when cAMP was omitted in the DNA binding reaction (Fig. 3, lane 4). Similarly, no retarded band was seen when a cell lysate of JM109(DE3) without pSK−CRP was used (Fig. 3, lane 3). Although the molecular mass of CRP is approximately 1.5 times (22 versus 14 kDa) that of InsA (2, 27), it generated a retarded band which migrated to the same level as that generated by InsA (Fig. 3, lanes 2, 5, and 6). A possible reason is that InsA may have the ability to bend DNA as CRP does, which would make the migration of DNA bands disproportional to the size of the bound protein. The same cell lysate containing CRP also generated retarded bands (Fig. 3, lane 10) in the presence of 20 mM cAMP when reacted with PlacZ, which is a 277-bp EcoRI-PvuII fragment containing the lacZ gene promoter, but did not generate any retarded bands with the 170-bp EcoRI-PvuII fragment of pBluescript II SK(+) which did not contain a CRP binding site (Fig. 3, lane 8).
FIG. 3.
Binding of the cAMP-CRP complex to the major IS2 promoter. PinsA is an 89-bp EcoRI-SpeI DNA fragment containing the major IS2 promoter, PlacZ is a 277-bp EcoRI-PvuII fragment containing the lac promoter, and NS is the 170-bp EcoRI-PvuII fragment of pBluescript II SK(+) which does not contain a CRP binding site. Three kinds of cell lysate were used: CRP, E. coli JM109(DE3) containing pSK−CRP; C, JM109(DE3) containing pT7-7; and InsA, JM109(DE3) containing pT7insA. The amounts of cell lysates used for each reaction with (+) or without (−) cAMP are as indicated. Solid arrows indicate bands of free DNA fragments, and open arrows indicate those of protein-bound DNA fragments.
The binding sequence of the cAMP-CRP complex on the major IS2 promoter.
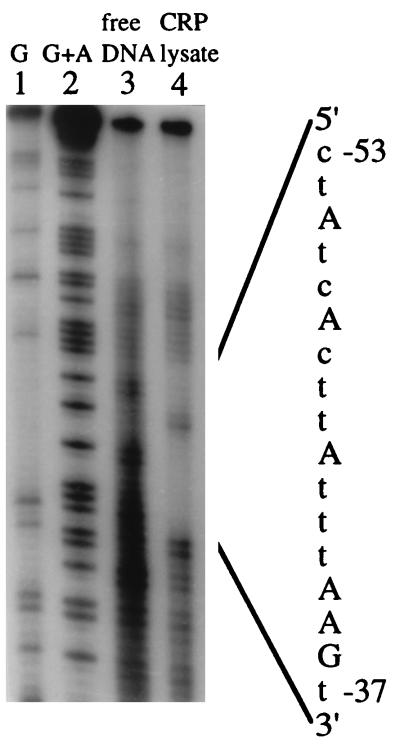
The DNA footprinting experiment was performed to determine the binding sequence of the cAMP-CRP complex on the major IS2 promoter. The same DNA fragment, PinsA, used for the gel retardation assay was used. This 89-bp EcoRI-BamHI fragment containing the IS2 LIR region was labeled at the 5′ EcoRI end and then incubated with the cAMP-CRP complex before footprinting. The same fragment was also subjected to Maxam-Gilbert sequencing reactions (44). All the reaction mixtures were electrophoresed on a 5% DNA sequencing gel. A footprint was seen on DNA reacted with the cAMP-CRP complex when the gel was autoradiographed (Fig. 4). The binding sequence of the cAMP-CRP complex was deduced to be 5′-CTATCACTTATTTAAGT-3′ (Fig. 4) by comparing the footprinting patterns of PinsA incubated with (Fig. 4, lane 4) or without (Fig. 4, lane 3) cAMP-CRP complex with the banding patterns of the Maxam-Gilbert G (Fig. 4, lane 1) and G+A (Fig. 4, lane 2) sequencing reactions. This sequence is complementary to the sequence 5′-ACTTAAATAAGTGATAG-3′ (IS2 nucleotide positions 37 to 53), which is located on the upper strand of the sequence shown in Fig. 1.
FIG. 4.
Determination of the cAMP-CRP complex binding sequence in the major IS2 promoter. The 89-bp EcoRI-SpeI DNA fragment containing the cAMP-CRP binding site was labeled by the Klenow enzyme with [α-32P]dATP at the EcoRI end, incubated with the cell lysate containing cAMP-CRP, and subjected to in situ DNA footprinting. Lanes 1 and 2, Maxam-Gilbert (44) G and G+A reactions, respectively, of the DNA fragment; lane 3, footprinting reaction of unbound DNA; lane 4, footprinting reaction of the cAMP-CRP-bound DNA. The binding sequence of the cAMP-CRP complex is deduced from the footprint as shown. The uppercase letters on the right represent sequences actually determined from the gel; the lowercase letters were filled in on the basis of known sequences.
Effect of cAMP-CRP on the transcription of the major IS2 promoter.
To determine whether the cAMP-CRP complex has any effect on transcription from the major IS2 promoter, the major IS2 promoter was fused with a promoterless luciferase gene and assayed for transcription in the presence or absence of cAMP-CRP. The 6.8-kb SalI-ScaI fragment containing the IS2 promoter fused with the lacZ structural gene was isolated from pInsApLacZ (27) and then ligated with the 3.2-kb SalI-HincII fragment containing the p15A replication origin and the chloramphenicol resistance gene of pACYC184, resulting in pACPIS2-LacZ (Fig. 2B). To replace the lacZ gene with the luciferase gene luxAB1 (45), the plasmid pACPIS2-LacZ was digested with ClaI. The ClaI ends were filled in with the Klenow enzyme, and the fragment was further digested with HindIII to delete the lacZ gene, producing a 3.2-kb DNA fragment. This 3.2-kb fragment was then ligated with the 2.3-kb HindIII-Ecl136II DNA fragment containing the luxAB1 (45) gene from pUCD1752 (a gift from C. I. Kado), generating pACPIS2-Lux (Fig. 2C).
The plasmid pACPIS2-Lux was introduced into the isogenic E. coli strains TP7811, TP7839, and TP7860, and the transformed cells were then assayed for the production of luciferase. TP7811 is wild type for cAMP-CRP, TP7839 is defective for CRP, and TP7860 is unable to produce cAMP. The major IS2 promoter was found to be able to drive the expression of the luciferase gene in TP7811 and produced 2.7 × 105 RLU of luciferin (Table 1). However, a fourfold increase (10.8 × 105 RLU) in the production of luciferase was seen when the same plasmid was introduced into the CRP− TP7839. A more profound (17.1-fold) increase in the production of luciferase was observed when pACPIS2-Lux was introduced into the Cya− TP7860. To ensure that the difference in the expression of the luciferase gene was not due to a difference in the copy number of pACPIS2-Lux in different hosts, plasmid DNA was isolated from the same numbers of TP7811, TP7839, and TP7860 cells and then quantitated. No difference in the copy number of pACPIS2-Lux in TP7811, TP7839, or TP7860 was observed. These results indicate that the major IS2 promoter is more active in the absence of CRP or cAMP and suggest that the cAMP-CRP complex is a negative regulator of the major IS2 promoter.
TABLE 1.
Effect of CRP and cAMP on transcription from the major IS2 promoter
| Bacterial strain | Luciferase activity (RLU)a | Relative fold increaseb |
|---|---|---|
| TP7811 (WT)c | (2.7 ± 0.5) × 105 | 1.0 |
| TP7839 (Δcrp) | (10.8 ± 0.6) × 105 | 4.0 |
| TP7860 (Δcya) | (46.3 ± 3.0) × 105 | 17.1 |
Each value represents the mean of four independent assays ± standard deviation.
Fold increase in RLU is relative to that of TP7811(pACPIS2-Lux), which is set as 1.
WT, wild type.
Effect of cAMP-CRP on IS2 transposition.
Since the major IS2 promoter is responsible for the transcription of the IS2 transposase InsAB′, the effect of cAMP-CRP on IS2 transposition was examined. A plasmid containing a mini-IS2 with a kanamycin resistance marker was constructed as follows. The plasmid pKS+ISF (27) was digested with AccI and HpaI to delete the IS2 sequence from nucleotide 578 to 1173, resulting in the plasmid pKS+ISFd1. IS2 nucleotides 103 to 440 were then removed by deleting the 337-bp XhoI-SmaI fragment of pKS+ISFd1. The 1.3-kb HincII fragment containing a kanamycin resistance gene from pUC4K (Pharmacia Biotech, Uppsala, Sweden) was then inserted into the blunt-ended XhoI and SmaI sites of pKS+ISFd1, generating pMIS2K (Fig. 2D). Since a total of 937 bp of internal IS2 sequence in pMIS2K were deleted, all functional genes of IS2 were destroyed and the mini-IS2 in this plasmid could transpose only in hosts that harbor IS2.
To ensure that strains TP7811, TP7839, and TP7860 contained IS2, PCR was performed with primers IS700–720 (5′-ATGCGCCAGAATGCGCTGTTG-3′) and IS1294–1273 (5′-TTAACCCATTACAAGCCCGCTG-3′), which amplify IS2 from nucleotide 700 to 1294. An expected 595-bp fragment was amplified from all three hosts. This fragment was sequenced, and the sequence was verified to be derived from IS2. pMIS2K was then introduced into TP7811, TP7839, and TP7860 containing pCJ105. The frequency of IS2 transposition onto pCJ105 in each host was then determined by mating pCJ105 out to another host. The results of this experiment are summarized in Table 2. In the wild-type host (TP7811), IS2 transposed at a frequency of 10−5. A 130-fold increase in transposition frequency was observed in the CRP− host, TP7839. A much higher transposition frequency (290-fold increase) was seen in the Cya− host, TP7860. These results indicate that IS2 transposes more efficiently in hosts defective in CRP or adenyl cyclase, suggesting that IS2 transposition is negatively regulated by the cAMP-CRP complex.
TABLE 2.
Effect of the cAMP-CRP complex on IS2 transposition
| Bacterial strain | Transposition frequencya | Relative fold increaseb |
|---|---|---|
| TP7811 (WT)c | (1.0 ± 0.3) × 10−5 | 1.0 |
| TP7839 (Δcrp) | (1.3 ± 0.5) × 10−3 | 130 |
| TP7860 (Δcya) | (2.9 ± 0.1) × 10−3 | 290 |
Numbers represent transposition frequencies of IS2 in different hosts and are the means (± standard deviations) of four independent experiments.
Fold increase in transposition frequency is relative to that of the mini-IS2 in TP7811, which is set as 1.
WT, wild type.
DISCUSSION
The consensus binding sequence for the cAMP-CRP complex is 5′-AAnTGTGAnnTnnnnCAnATT-3′ (11). This sequence is found in the major IS2 promoter at three regions: IS2 nucleotide positions 34 to 54, 43 to 63, and 42 to 22 (Fig. 1). The sequence of region III (IS2 nucleotides 42 to 22) is located on the lower strand of the IS2 LIR, whereas those of region I (IS2 nucleotides 34 to 54) and region II (IS2 nucleotides 43 to 63) are on the upper strand. Seven residues in region I, eight in region II, and six in region III conform to this 13-residue consensus cAMP-CRP binding sequence. This finding suggests that IS2 transposition may be subject to cAMP-CRP regulation. This hypothesis is supported by the demonstration that the cAMP-CRP complex binds to the major IS2 promoter (Fig. 3 and 4) and that binding of the cAMP-CRP complex to the IS2 promoter has a negative effect on transcription from this promoter (Table 1). As a consequence, the production of transposase is decreased and IS2 transposition frequency is reduced. This supposition was demonstrated in this study by the observation that IS2 transposition frequency is higher in E. coli mutants defective in CRP or adenyl cyclase than in a wild-type host (Table 2). It is also possible that the binding of the cAMP-CRP complex to the LIR interferes with the binding of the IS2 transposase to the same region to initiate transposition. This possibility remains to be investigated.
The cAMP-CRP complex was determined to bind the sequence 5′-ACTTAAATAAGTGATAG-3′ located at IS2 nucleotides 37 to 53 (Fig. 1). This sequence is located within region I of the three putative cAMP-CRP binding sites mentioned above. This cAMP-CRP binding sequence overlaps almost entirely with that of the InsA binding sequence, which is located at IS2 nucleotides 33 to 52 (Fig. 1). This area covers the entire −10 sequence and its flanking regions of the major IS2 promoter. We have previously shown that the binding of InsA to this region also suppresses transcription from this promoter and thus decreases IS2 transposition frequency (27). In this study, we have demonstrated that the cAMP-CRP complex binds to the same region and has the same suppressive effect as the InsA on IS2 transposition. InsA is a native IS2 protein, whereas CRP is a host protein. It is conceivable that the host has a mechanism to limit IS2 transposition, since overtransposition could be detrimental to the host, but it is quite intriguing to find that IS2 produces a protein to suppress its own transposition. It remains to be investigated whether InsA and the cAMP-CRP complex compete with each other for binding to the same site. It appears that InsA and the cAMP-CRP complex do not bind concomitantly, since a cell lysate containing both proteins did not produce a band that migrated more slowly than the one produced by either InsA or the cAMP-CRP complex alone in the gel retardation assay (data not shown). It is unknown whether there is a mechanism to coordinate the binding of these two negative regulators. How IS2 derepresses the suppression by InsA or the cAMP-CRP complex also remains to be studied.
The cAMP-CRP complex of E. coli is involved in the activation or repression (34, 50, 62, 63) of many genes. For example, the cAMP-CRP complex alone activates the transcription of lacp1 (42), galp1 (48, 49), malTp (67), PC and PBAD of the AraCBAD operon (40), papp (15), and gyrA (19). In the absence of the CytR repressor, it also activates deop2 (46, 53, 72), tsx-p2 (17), nupG (47), and cdd (25). The promoters malKp and malEp (62, 67) require both the MalT and the cAMP-CRP complexes, and that of the ansB gene (31) requires both the FNR and the cAMP-CRP complexes for transcriptional activation.
In this study, the binding of the cAMP-CRP complex was found to have a negative effect on transcription from the major IS2 promoter. Negative regulation by the cAMP-CRP complex has also been demonstrated to occur on lacp2/p3 (13, 42, 79), galp2 (48), proPp1 (80), and crp (1, 30). In addition, the cAMP-CRP complex together with the CytR repressor inhibits the transcription of deop2 (46, 53, 58, 72), cytRp (23, 52), tsx-p2 (17), nupG (51), and cdd (25).
The effect of cAMP-CRP on IS2 transposition described in this report is the first example of regulation of the transposition of transposable elements by the cAMP-CRP complex. It is not known whether the cAMP-CRP complex has a similar effect on other members of the IS3 family. We have searched for the presence of the cAMP-CRP binding sequence on the transposable elements of the IS3 family (56, 65, 66) and found that 9 of 11 members of the IS3 family have a putative CRP binding sequence on the promoter region (Table 3) which has 7 or more bp matched with the 13-bp consensus sequence, suggesting that the cAMP-CRP complex also regulates their transpositions. We also found that 11 of 13 non-IS3 transposable elements have 7 or more bp that matched with the putative CRP binding sequence (Table 4). Whether the cAMP-CRP complex has any effect on the transposition of these transposable elements remains to be determined.
TABLE 3.
Putative cAMP-CRP binding sequences present in members of the IS3 family
| TEa | Promoter | Sequenceb | Nucleotide positions | Matchc | Reference(s) |
|---|---|---|---|---|---|
| IS3411 | orfV | AATCGTGAACTGCGCCGCAGT | 292–312 | 10 | 29 |
| IS51 | orf1 | ACCGGTGGAATATGACCGATT | 104–124 | 9 | 81 |
| IS476 | orf1 | AGTTGTGCCGTCGGCATGGTT | 141–161 | 8 | 32 |
| IS3 | orfA | TCATGTGAGTCACCTCTGACT | 69–49 | 8 | 75 |
| IS911 | orfA | CACACTGAATTTGGCCACCTG | 9–29 | 8 | 56 |
| IS150 | orfA | AAATGGAATAGCCCCTAATAT | 29–49 | 7 | 66 |
| IS186 | orf1 | GGTTGTGGTATTACGCCTGAT | 18–38 | 7 | 9, 70 |
| IS600 | orfA | CCTTCTGATGCCATTCTATTT | 56–36 | 7 | 43 |
| IS861 | orf1 | ATAACTTAATTTCATCAGAAA | 128–108 | 7 | 65 |
TABLE 4.
Putative cAMP-CRP binding sequences present in non-IS3 family transposable elements
| TEa | Promoter | Sequenceb | Nucleotide positions | Matchc | Reference(s) |
|---|---|---|---|---|---|
| Tn1000 | tnpR | AAATGTATCCTAAATCAAATA | 2779–2799 | 10 | 6 |
| Tn1000 | tnpA | ATGTGTGCGATAATTTATAAT | 2865–2885 | 9 | 6 |
| IS4 | tnp | ACAAGTGAGCGTTTCCGGATT | 70–50 | 9 | 33, 77 |
| IS30 | orfA | AACTGTTGCGTTGACCAATTG | 29–9 | 9 | 10 |
| Tn3 | tnpR | AAATGTACCTTAAATCGAATA | 3207–3187 | 9 | 24 |
| Tn3 | tnpA | CTATGTCTGATAATTTATAAT | 3121–3101 | 7 | 24 |
| IS15 | orfI | GATGGTGGCGTAAGCCGTCTT | 409–429 | 8 | 76 |
| IS50 | tnp | TATCATGAACGTTACCATGTT | 98–78 | 8 | 35 |
| Tn7 | tnsA | CAGTATGCTTTTTCACAGCAT | 124–104 | 8 | 14 |
| IS102 | orfI | TCTGGTGATTAAACGCGTATT | 254–274 | 8 | 3 |
| IS21 | istA | GATTGTCCGCTCCACCCAACA | 40–60 | 7 | 59 |
| IS10 | pIN | AGATGTGTATCCACCTTAACT | 58–38 | 7 | 22, 71 |
| IS10 | pOUT | AGATGTGCGAACTCGATATTT | 106–126 | 7 | 22, 71 |
| IS903 | tnp | ATGCGTGATCTGATCCTTCAA | 1005–1025 | 7 | 21 |
ACKNOWLEDGMENTS
We thank C.-H. Lee for critical reading and editing of the manuscript, S.-C. Lo and S.-T. Liu for helpful comments on this work, and S. Tarbor and H. Aiba for providing plasmids and host cells.
This work was supported by grant NSC86-2314-B-010-032 from the National Science Council, Taiwan, R.O.C., to S.-T. Hu.
REFERENCES
- 1.Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983;32:141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- 2.Aiba H, Fujimoto S, Ozaki N. Molecular cloning and nucleotide sequencing of the gene for E. coli cAMP receptor protein. Nucleic Acids Res. 1982;10:1345–1361. doi: 10.1093/nar/10.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardi A, Bernardi F. Complete sequence of an IS element present in pSC101. Nucleic Acids Res. 1981;9:2905–2911. doi: 10.1093/nar/9.12.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biville F, Blazy B, Guiso N. Transcription termination factor Rho of Escherichia coli K-12: some regulatory aspects of its expression and activity. Biochimie. 1983;65:339–344. doi: 10.1016/s0300-9084(83)80155-2. [DOI] [PubMed] [Google Scholar]
- 5.Boyer H W, Roulland-Dusspix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 6.Broom J E, Hill D F, Hughes G, Jones W A, McNaughton J C, Stockwell P A, Petersen G B. Sequence of a transposon identified as Tn1000 (gamma delta) DNA Sequence. 1995;5:185–189. doi: 10.3109/10425179509029361. [DOI] [PubMed] [Google Scholar]
- 7.Bullock W O, Ferrnandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechnology. 1987;4:376–379. [Google Scholar]
- 8.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 9.Chong P, Hui I, Loo T, Gillam S. Structural analysis of a new GC-specific insertion element IS186. FEBS Lett. 1985;192:47–52. doi: 10.1016/0014-5793(85)80040-5. [DOI] [PubMed] [Google Scholar]
- 10.Dalrymple B, Caspers P, Arber W. Nucleotide sequence of the prokaryotic mobile genetic element IS30. EMBO J. 1984;3:2145–2149. doi: 10.1002/j.1460-2075.1984.tb02104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Crombrugghe B, Busby S, Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984;224:831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- 12.Engler J A, van Bree M P. The nucleotide sequence and protein-coding capability of the transposable element IS5. Gene. 1981;14:155–163. doi: 10.1016/0378-1119(81)90111-6. [DOI] [PubMed] [Google Scholar]
- 13.Eschenlauer A C, Reznikoff W S. Escherichia coli catabolite gene activator protein mutants defective in positive control of lac operon transcription. J Bacteriol. 1991;173:5024–5029. doi: 10.1128/jb.173.16.5024-5029.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores C, Qadri M I, Lichtenstein C. DNA sequence analysis of five genes; tnsA, B, C, D, and E, required for Tn7 transposition. Nucleic Acids Res. 1990;18:901–911. doi: 10.1093/nar/18.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsman K, Sonden B, Goransson M, Uhlin B E. Antirepression function in Escherichia coli for the cAMP-cAMP receptor protein transcriptional activator. Proc Natl Acad Sci USA. 1992;89:9880–9884. doi: 10.1073/pnas.89.20.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner M M, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlach P, Søgaard-Andersen L, Pedersen H, Martinussen J, Valentin-Hansen P, Bremer E. The cyclic AMP (cAMP)-cAMP receptor protein complex functions both as an activator and as a corepressor at the tsx-p2 promoter of Escherichia coli K-12. J Bacteriol. 1991;173:5419–5430. doi: 10.1128/jb.173.17.5419-5430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosal D, Sommer H, Saedler H. Nucleotide sequence of the transposable DNA-element IS2. Nucleic Acids Res. 1979;6:1111–1121. doi: 10.1093/nar/6.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Gomez J M, Baquero F, Blazquez J. Cyclic AMP receptor protein positively controls gyrA transcription and alters DNA topology after nutritional upshift in Escherichia coli. J Bacteriol. 1996;178:3331–3334. doi: 10.1128/jb.178.11.3331-3334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grindley N D F, Leschziner A E. DNA transposition: from a black box to a colour monitor. Cell. 1995;83:1063–1066. doi: 10.1016/0092-8674(95)90132-9. [DOI] [PubMed] [Google Scholar]
- 21.Grindley N D F, Joyce C M. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harbor Symp Quant Biol. 1981;45:125–133. doi: 10.1101/sqb.1981.045.01.021. [DOI] [PubMed] [Google Scholar]
- 22.Halling S M, Simons R W, Way J C, Walsh R B, Kleckner N. DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. Proc Natl Acad Sci USA. 1982;79:2608–2612. doi: 10.1073/pnas.79.8.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hans-Henrik K, Valentin-Hansen P, Søgaard-Anderson L. CytR/cAMP-CRP nucleoprotein formation in E. coli: the CytR repressor binds its operator as a stable dimer in a ternary complex with cAMP-CRP. J Mol Biol. 1996;260:113–119. doi: 10.1006/jmbi.1996.0385. [DOI] [PubMed] [Google Scholar]
- 24.Heffron F, McCarthy B J. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979;18:1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- 25.Holst B, Søgaard-Andersen L, Pedersen H, Valentin-Hansen P. The cAMP-CRP/CytR nucleoprotein complex in Escherichia coli: two pairs of closely linked binding sites for the cAMP-CRP activator complex are involved in combinatorial regulation of the cdd promoter. EMBO J. 1992;11:3635–3643. doi: 10.1002/j.1460-2075.1992.tb05448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu S T, Lee C H. Characterization of the transposon carrying the STII gene of enterotoxigenic Escherichia coli. Mol Gen Genet. 1988;214:490–495. doi: 10.1007/BF00330485. [DOI] [PubMed] [Google Scholar]
- 27.Hu S T, Hwang J H, Lee L C, Lee C H, Li P L, Hsieh Y C. Functional analysis of the 14 kDa protein of insertion sequence 2. J Mol Biol. 1994;236:503–513. doi: 10.1006/jmbi.1994.1161. [DOI] [PubMed] [Google Scholar]
- 28.Hu S T, Lee L C, Lei G S. Detection of an IS2-encoded 46-kDa protein capable of binding terminal repeats of IS2. J Bacteriol. 1996;178:5652–5659. doi: 10.1128/jb.178.19.5652-5659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishiguro N, Sato G. Nucleotide sequence of insertion sequence IS3411, which flanks the citrate utilization determinant of transposon Tn3411. J Bacteriol. 1988;170:1902–1906. doi: 10.1128/jb.170.4.1902-1906.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishizuka H, Hanamura A, Inada T, Aiba H. Mechanism of the down-regulation of cAMP receptor protein by glucose in Escherichia coli: role of autoregulation of the crp gene. EMBO J. 1994;13:3077–3082. doi: 10.1002/j.1460-2075.1994.tb06606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jennings M P, Beacham I R. Co-dependent positive regulation of the ansB promoter of Escherichia coli by CRP and the FNR protein: a molecular analysis. Mol Microbiol. 1993;9:155–164. doi: 10.1111/j.1365-2958.1993.tb01677.x. [DOI] [PubMed] [Google Scholar]
- 32.Kearney B, Staskawicz B J. Characterization of IS476 and its role in bacterial spot disease of tomato and pepper. J Bacteriol. 1990;172:143–148. doi: 10.1128/jb.172.1.143-148.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klaer R, Kuehn S, Tillmann E, Fritz H-J, Starlinger P. The sequence of IS4. Mol Gen Genet. 1981;181:169–175. doi: 10.1007/BF00268423. [DOI] [PubMed] [Google Scholar]
- 34.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 35.Krebs M P, Reznikoff W S. Transcriptional and translational inhibition sites of IS50: control of transposase and inhibitor expression. J Mol Biol. 1986;192:781–791. doi: 10.1016/0022-2836(86)90028-8. [DOI] [PubMed] [Google Scholar]
- 36.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuwabara M D, Sigman D S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987;26:7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- 38.Lei G S, Hu S T. Functional domains of the InsA protein of IS2. J Bacteriol. 1997;179:6238–6243. doi: 10.1128/jb.179.20.6238-6243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis L A, Grindley N D F. Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol Microbiol. 1997;25:517–529. doi: 10.1046/j.1365-2958.1997.4871848.x. [DOI] [PubMed] [Google Scholar]
- 40.Lobell R B, Schleif R F. AraC-DNA looping: orientation and distance-dependent loop breaking by the cyclic AMP receptor protein. J Mol Biol. 1991;218:45–54. doi: 10.1016/0022-2836(91)90872-4. [DOI] [PubMed] [Google Scholar]
- 41.Mahillon J, Seurinck J, Van Rompuy L, Delcour J, Zabeau M. Nucleotide sequence and structural organization of an insertion sequence element (IS231) from Bacillus thuringiensis strain verliner 1715. EMBO J. 1985;4:3895–3899. doi: 10.1002/j.1460-2075.1985.tb04163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malan T P, McClure W R. Dual promoter control of the Escherichia coli lactose operon. Cell. 1984;39:173–180. doi: 10.1016/0092-8674(84)90203-4. [DOI] [PubMed] [Google Scholar]
- 43.Matsutani S, Ohtsubo H, Maeda Y, Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987;196:445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- 44.Maxam A, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavage. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 45.Meighen E A. Enzymes and genes from the lux operons of bioluminescent bacteria. Annu Rev Microbiol. 1988;42:151–176. [Google Scholar]
- 46.Møllegaard N E, Rasmussen P B, Valentin-Hansen P, Nielsen P E. Characterization of promoter recognition complexes formed by CRP and CytR for repression and by CRP and RNA polymerase for activation of transcription on the Escherichia coli deoP2 promoter. J Biol Chem. 1993;268:17471–17477. [PubMed] [Google Scholar]
- 47.Munch-Petersen A, Jensen N. Analysis of the regulatory region of the Escherichia coli nupG gene, encoding a nucleoside-transport protein. Eur J Biochem. 1990;190:541–551. doi: 10.1111/j.1432-1033.1990.tb15608.x. [DOI] [PubMed] [Google Scholar]
- 48.Musso R E, Lauro R D, Adhya S, de Crombrugghe B. Dual control for transcription of the galactose operon by cyclic AMP and its receptor protein at two interspersed promoters. Cell. 1977;12:847–854. doi: 10.1016/0092-8674(77)90283-5. [DOI] [PubMed] [Google Scholar]
- 49.Nissley S P, Anderson W B, Gottesman M E, Perlman R L, Pastan I. In vitro transcription of the gal operon requires cyclic adenosine monophosphate and cyclic adenosine monophosphate receptor protein. J Biol Chem. 1971;246:4671–4678. [PubMed] [Google Scholar]
- 50.Pastan I, Perlman R L. Cyclic adenosine monophosphate in bacteria. Science. 1970;169:339–344. doi: 10.1126/science.169.3943.339. [DOI] [PubMed] [Google Scholar]
- 51.Pedersen H, Dall J, Dandanell G, Valentin-Hansen P. Gene-regulatory modules in Escherichia coli: nucleoprotein complexes formed by cAMP-CRP and CytR at the nupG promoter. Mol Microbiol. 1995;17:843–853. doi: 10.1111/j.1365-2958.1995.mmi_17050843.x. [DOI] [PubMed] [Google Scholar]
- 52.Pedersen H, Søgaard-Andersen L, Holst B, Gerlach P, Bremer E, Valentin-Hansen P. cAMP-CRP activator complex and the CytR repressor protein bind cooperatively to the cytRP promoter in Escherichia coli and CytR antagonizes the cAMP-CRP-induced DNA bend. J Mol Biol. 1992;227:396–406. doi: 10.1016/0022-2836(92)90896-r. [DOI] [PubMed] [Google Scholar]
- 53.Pedersen H, Søgaard-Andersen L, Holst B, Valentin-Hansen P. Heterologous cooperativity in Escherichia coli: the CytR repressor contacts both DNA and the cAMP receptor protein when binding to the deoP2 promoter. J Biol Chem. 1991;266:17804–17808. [PubMed] [Google Scholar]
- 54.Polard P, Chandler M. Bacterial transposases and retroviral integrase. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 55.Polard P, Prere M F, Chandler M, Fayet O. Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J Mol Biol. 1991;222:465–477. doi: 10.1016/0022-2836(91)90490-w. [DOI] [PubMed] [Google Scholar]
- 56.Prere M-F, Chandler M, Fayet O. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990;172:4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Priefer U B, Kalinowski J, Ruger B, Heumann W, Puhler A. ISR1, a transposable DNA sequence resident in Rhizobium class IV strains, shows structural characteristics of classical insertion elements. Plasmid. 1989;21:120–128. doi: 10.1016/0147-619x(89)90055-3. [DOI] [PubMed] [Google Scholar]
- 58.Rasmussen P B, Søgaard-Andersen L, Valentin-Hansen P. Identification of the nucleotide sequence recognized by the cAMP-CRP dependent CytR repressor protein in the deoP2 promoter in E. coli. Nucleic Acids Res. 1993;21:879–885. doi: 10.1093/nar/21.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reimmann C, Moore R, Little S, Savioz A, Willetts N S, Haas D. Genetic structure, function, and regulation of the transposable element IS21. Mol Gen Genet. 1989;215:416–424. doi: 10.1007/BF00427038. [DOI] [PubMed] [Google Scholar]
- 60.Rezsohazy R, Hallet B, Delcour J, Mahillon J. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 61.Rezsohazy R, Hallet B, Mahillon J, Delcour J. IS231 V and W from Bacillus thuringiensis subsp. israelensis, two distant members of the IS231 family of insertion sequences. Plasmid. 1993;30:141–149. doi: 10.1006/plas.1993.1041. [DOI] [PubMed] [Google Scholar]
- 62.Richet E, Vidal-Ingigliardi D, Raibaud O. A new mechanism for coactivation of transcription initiation: repositioning of an activator triggered by the binding of a second activator. Cell. 1991;66:1185–1195. doi: 10.1016/0092-8674(91)90041-v. [DOI] [PubMed] [Google Scholar]
- 63.Rickenberg H. Cyclic AMP in prokaryotes. Annu Rev Microbiol. 1974;28:353–369. doi: 10.1146/annurev.mi.28.100174.002033. [DOI] [PubMed] [Google Scholar]
- 64.Ronecker H J, Rak B. Genetic organization of insertion element IS2 based on a revised nucleotide sequence. Gene. 1987;59:291–296. doi: 10.1016/0378-1119(87)90337-4. [DOI] [PubMed] [Google Scholar]
- 65.Rubens C E, Heggen L M, Kuypers J M. IS861, a group B streptococcal insertion sequence related to IS150 and IS3 of Escherichia coli. J Bacteriol. 1989;171:5531–5535. doi: 10.1128/jb.171.10.5531-5535.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz E, Kroger M, Rak B. IS150: distribution, nucleotide sequence and phylogenetic relationships of a new E. coli insertion element. Nucleic Acids Res. 1988;16:6789–6802. doi: 10.1093/nar/16.14.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz M. The maltose regulon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1482–1502. [Google Scholar]
- 68.Sekine Y, Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci USA. 1989;86:4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- 70.Sengstag C, Iida S, Hiestand-Nauer R, Arber W. Terminal inverted repeats of prokaryotic transposable element IS186 which can generate duplications of variable length at an identical target sequence. Gene. 1986;49:153–156. doi: 10.1016/0378-1119(86)90395-1. [DOI] [PubMed] [Google Scholar]
- 71.Simons R W, Hoopes B C, McClure W R, Kleckner N. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell. 1983;34:673–682. doi: 10.1016/0092-8674(83)90400-2. [DOI] [PubMed] [Google Scholar]
- 72.Søgaard-Andersen L, Møllegaard N E, Douthwaite S R, Valentin-Hansen P. Tandem DNA-bound cAMP-CRP complexes are required for transcriptional repression of the deoP2 promoter by the CytR repressor in Escherichia coli. Mol Microbiol. 1990;4:1595–1601. [PubMed] [Google Scholar]
- 73.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of a cloned gene. J Biol Chem. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 74.Tabor, S. Personal communication.
- 75.Timmerman K P, Tu C-P D. Complete sequence of IS3. Nucleic Acids Res. 1985;13:2127–2139. doi: 10.1093/nar/13.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the transposable element IS15. Gene. 1984;30:113–120. doi: 10.1016/0378-1119(84)90111-2. [DOI] [PubMed] [Google Scholar]
- 77.Trinks K, Habermann P, Beyreuther K, Starlinger P, Ehring R. An IS4-encoded protein is synthesized in minicells. Mol Gen Genet. 1981;182:183–188. doi: 10.1007/BF00269656. [DOI] [PubMed] [Google Scholar]
- 78.Vogele K, Schwartz E, Welz C, Schiltz E, Rak B. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 1991;19:4377–4385. doi: 10.1093/nar/19.16.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiong X, de la Cruz N, Reznikoff W S. Downstream deletion analysis of the lac promoter. J Bacteriol. 1991;173:4570–4577. doi: 10.1128/jb.173.15.4570-4577.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu J, Johnson R C. Cyclic AMP receptor protein functions as a repressor of the osmotically inducible promoter proP P1 in Escherichia coli. J Bacteriol. 1997;179:2410–2417. doi: 10.1128/jb.179.7.2410-2417.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamada T, Lee P-D, Kosuge T. Insertion sequence elements of Pseudomonas savastanoi: nucleotide sequence and homology with Agrobacterium tumefaciens transfer DNA. Proc Natl Acad Sci USA. 1986;83:8263–8267. doi: 10.1073/pnas.83.21.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]