Figure 2. DDX39A is antiviral independent of the interferon pathway.
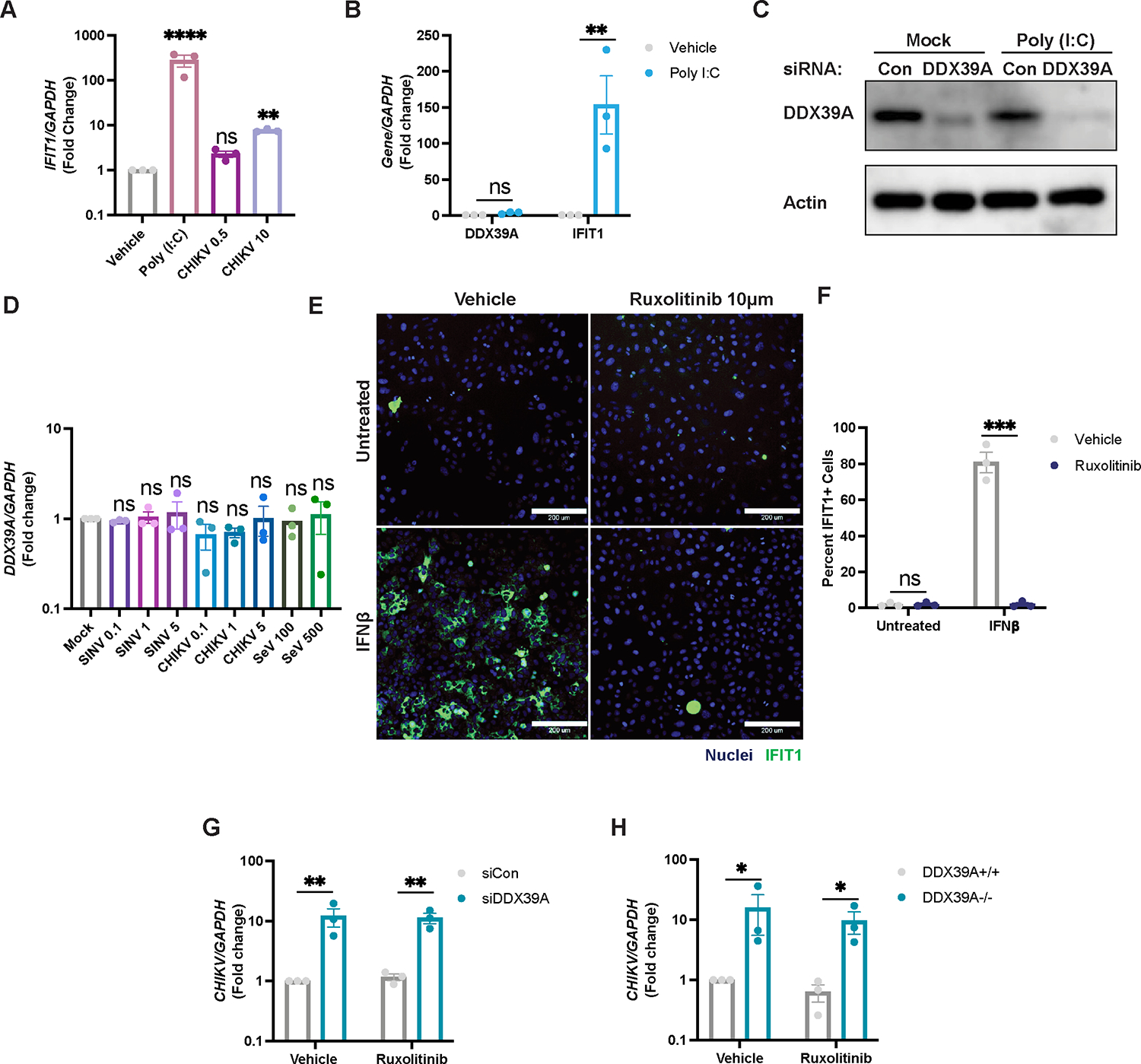
(A) U2OS cells were stimulated with poly (I:C) or infected with CHIKV at the indicated MOIs. IFIT1 levels were assessed by qPCR relative to uninfected vehicle control. (B-C) Levels of DDX39A or IFIT1 were measured by (B) qPCR for mRNA levels and by (C) immunoblot. A representative immunoblot image is shown. (D) U2OS cells were infected with SINV-mKate (MOIs 0.1,1,5), CHIKV (MOIs 0.1,1,3) or SeV (100 and 500 HAU/mL), and levels of DDX39A were assessed by qPCR compared to mock uninfected cells (E) Representative immunofluorescence image of U2OS cells pretreated with vehicle or Ruxolitinib (10 μM) for 2h and stimulated with IFNβ (10 ng/mL) for 6h, and immunostained (nuclei, Hoechst 33342 in blue and IFIT1+ cells in green). Magnification 10x; Scale bar 200 μm. (F) Percent IFIT1+ cells were calculated with automated microscopy. (G) Control or siRNA depleted-DDX39A, and (H) DDX39A+/+ or DDX39A−/− U2OS cells were pretreated with vehicle or Ruxolitinib (10 μM) and infected with CHIKV (MOI 0.5, 24h). Levels of viral RNA were measured by qPCR. Data are presented as fold change vs. vehicle-treated cells (A, B, F, G, H). Dots represent individual knockdown experiments. Immunoblots were performed in duplicate, n=3 for all other experiments as indicated. GAPDH was used as a loading control gene for all qPCR experiments. Beta-Actin was used as a loading control for all immunoblots as indicated. Statistical analyses were performed using one-way ANOVA with Bonferroni corrections (A, D), two-way ANOVA with Bonferroni corrections for multiple comparisons (B, E, G, H) *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; error bars represent S.E.M.
