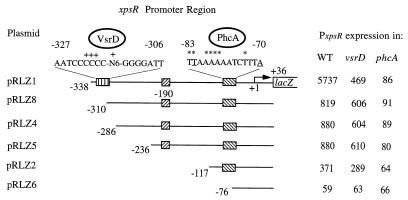
FIG. 2.
Identification of PxpsR sequences required for transcriptional regulation by VsrD and PhcA. The xpsR promoter and various lengths of upstream sequences were fused to lacZ on pRG970 to generate plasmids pRLZ1 to pRLZ8. PxpsR expression (i.e., transcription directed by each PxpsR fragment) was monitored in the wild type (WT), strain AW-D5 (vsrD mutant), and strain AW1-80 (phcA mutant) of R. solanacearum by measuring LacZ activity (β-galactosidase activity in Miller units) as described in Table 2, footnote a. Nucleotide numbering is relative to the transcription start site of xpsR (+1|→). Striped and hatched boxes and associated sequences illustrate putative binding sites for VsrD and PhcA, respectively. The symbol + above the putative VsrD binding site dyad indicates the positions of mutations that eliminated VsrD activation of PxpsR.; N6 represents the sequence TAAATT. Underlined nucleotides in the PhcA binding site indicate the T-N11-A motif found in the binding sites of nearly all LysR-type activators (31). Asterisks indicate the positions of mutations that affected PhcA binding and activation of PxpsR (Table 3). The complete xpsR promoter region sequence is GenBank under accession no. U18136. pRLZ7 (Table 1) had PxpsR activity similar to that of pRLZ1.