Extended Data Figure 2.
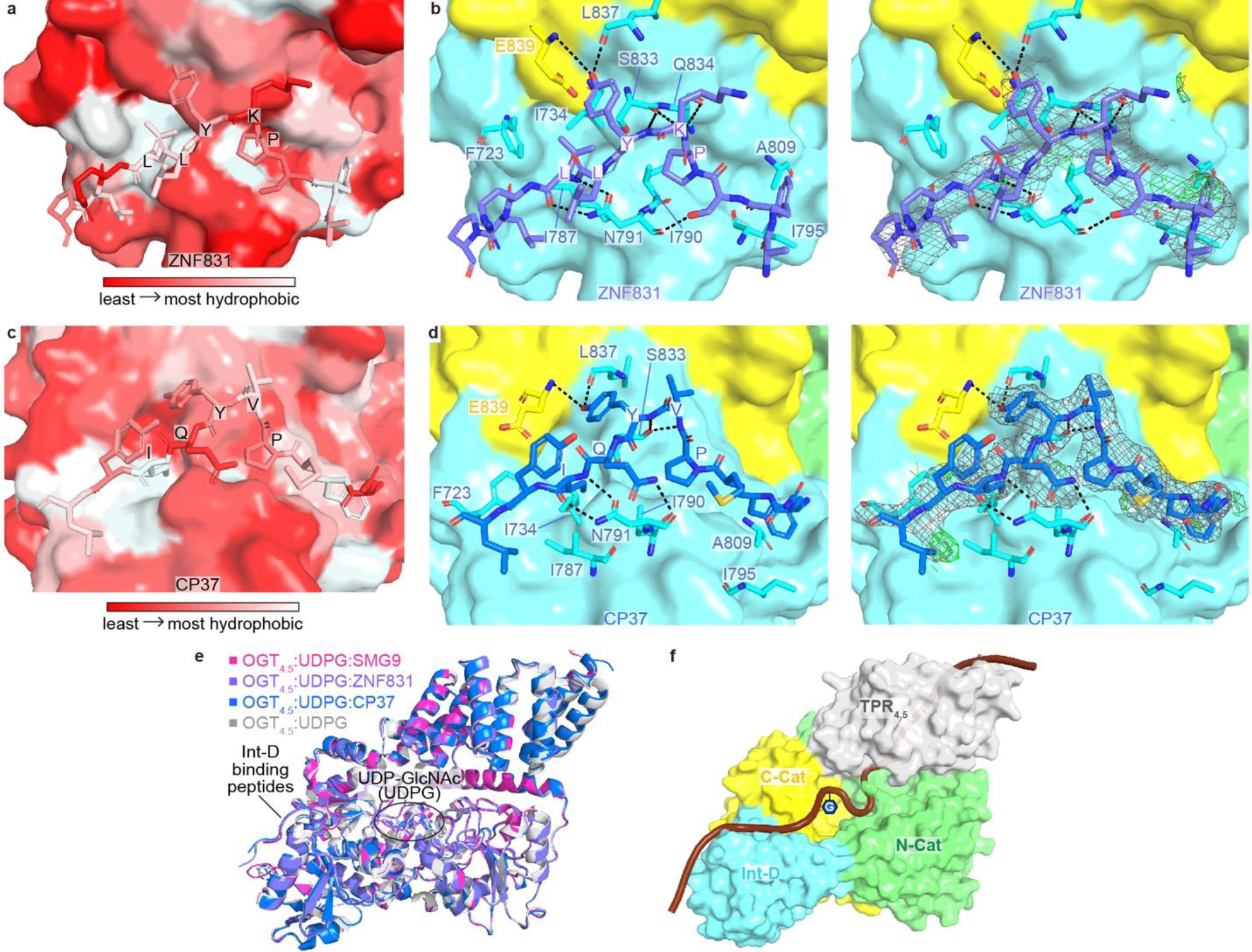
(a) Zoom-in view at the Int-D binding site demonstrating hydrophobic interactions between ZNF831 peptide (shown as sticks) and Int-D in OGT4.5 (shown as surface). Red and white colors represent the least and most hydrophobic areas, respectively. (b) Zoom-in view at the Int-D binding site demonstrating polar interactions between ZNF831 peptide (shown as purple sticks) and Int-D in OGT4.5 (shown as surface) with interacting OGT residues shown in sticks. 2Fo–Fc electron density map of ZNF831 peptide is shown as grey mesh and contoured at 1.0 σ, Fo–Fc electron density map is shown as green mesh and contoured at 3.0 σ. (c) Zoom-in view at the Int-D binding site demonstrating hydrophobic interactions between CP37 peptide (shown as sticks) and Int-D in OGT4.5 (shown as surface). Red and white colors represent the least and most hydrophobic areas, respectively. (d) Zoom-in view at the Int-D binding site demonstrating polar interactions between CP37 peptide (shown as blue sticks) and Int-D in OGT4.5 (shown as surface) with interacting OGT residues shown in sticks. 2Fo–Fc electron density map of CP37 peptide is shown as grey mesh and contoured at 1.0 σ, Fo–Fc electron density map is shown as green mesh and contoured at 3.0 σ. (e) Superimposition of OGT4.5:UDP-GlcNAc (4GZ5, grey), OGT4.5:UDP-GlcNAc:SMG9 (magenta), OGT4.5:UDP-GlcNAc:ZNF831 (purple), and OGT4.5:UDP-GlcNAc:CP37 (blue) crystal structures showing peptide binding to Int-D does not change the overall structure of OGT4.5. (f) A proposed model of a substrate peptide (shown as brown cartoon) binding to OGT4.5 (shown as surface, domains colored as in Figure 1a). Int-D and TPR domain interactions facilitate substate glycosylation in the active site of OGT. G symbol represents GlcNAc moiety.
